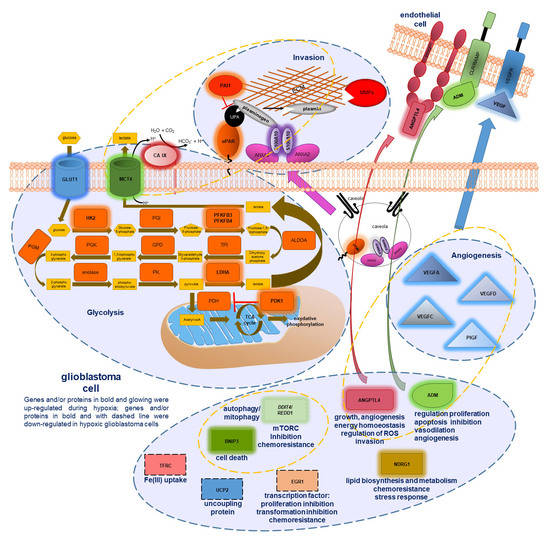
Investigating Glioblastoma Response to Hypoxia
Abstract
:1. Introduction
2. Experimental Section
2.1. Cell Lines and Cell Culture
2.2. Antibodies
2.3. Hypoxia Signaling Pathway RT2 PCR Profiler Array
2.4. RT-qPCR
2.5. Western Blotting
2.6. Statistical Analysis
2.7. Human GB Transcriptome Data
3. Results
3.1. Expression of Hypoxia Related Genes in Glioblastoma Cells
3.1.1. Transcription Factors Gene Expression Analysis
3.1.2. Expression Analysis of Glycolysis Related Genes
3.1.3. Expression Analysis of Angiogenic Genes
3.1.4. Expression Analysis of Invasion Genes
3.1.5. Expression Analysis of Other Hypoxia Related Genes
3.2. Expression of Hypoxia Related Proteins in GB Cells
3.3. Correlation of Hypoxia Altered Genes with Clinical Outcomes in GB Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J. Clin. Oncol. 2013, 31, 4085–4091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Wang, M.; Dignam, J.J.; Won, M.; Curran, W.; Mehta, M.; Gilbert, M.R. Variation over time and interdependence between disease progression and death among patients with glioblastoma on RTOG 0525. Neuro Oncol. 2015, 17, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.R.; Hill, R.; Pilkington, G.J.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Invasion. Cells 2017, 6, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castaldo, S.A.; Freitas, J.R.; Conchinha, N.V.; Madureira, P.A. The Tumorigenic Roles of the Cellular REDOX Regulatory Systems. Oxid. Med. Cell. Longev. 2016, 2016, 8413032. [Google Scholar] [CrossRef] [Green Version]
- Madureira, P.A.; Surette, A.P.; Phipps, K.D.; Taboski, M.A.; Miller, V.A.; Waisman, D.M. The role of the annexin A2 heterotetramer in vascular fibrinolysis. Blood 2011, 118, 4789–4797. [Google Scholar] [CrossRef] [Green Version]
- Madureira, P.A.; O’Connell, P.A.; Surette, A.P.; Miller, V.A.; Waisman, D.M. The biochemistry and regulation of S100A10: A multifunctional plasminogen receptor involved in oncogenesis. J. Biomed. Biotechnol. 2012, 2012, 353687. [Google Scholar] [CrossRef] [Green Version]
- Miller, V.A.; Madureira, P.A.; Kamaludin, A.A.; Komar, J.; Sharma, V.; Sahni, G.; Thelwell, C.; Longstaff, C.; Waisman, D.M. Mechanism of plasmin generation by S100A10. Thromb. Haemost. 2017, 117. [Google Scholar] [CrossRef]
- Gonzalez-Avila, G.; Sommer, B.; García-Hernández, A.A.; Ramos, C. Matrix Metalloproteinases’ Role in Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 97–131. [Google Scholar] [CrossRef]
- Howarth, A.; Simms, C.; Kerai, N.; Allen, O.; Mihajluk, K.; Madureira, P.A.; Sokratous, G.; Cragg, S.; Lee, S.Y.; Morley, A.D.; et al. DIVERSet JAG Compounds Inhibit Topoisomerase II and Are Effective Against Adult and Pediatric High-Grade Gliomas. Transl. Oncol. 2019, 12, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Mihajluk, K.; Simms, C.; Reay, M.; Madureira, P.A.; Howarth, A.; Murray, P.; Nasser, S.; Duckworth, C.A.; Pritchard, D.M.; Pilkington, G.J.; et al. IP1867B suppresses the insulin-like growth factor 1 receptor (IGF1R)ablating epidermal growth factor receptor inhibitor resistance in adult high grade gliomas. Cancer Lett. 2019, 458, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, S.A.; Ajime, T.; Serrão, G.; Anastácio, F.; Rosa, J.T.; Giacomantonio, C.A.; Howarth, A.; Hill, R.; Madureira, P.A. Annexin A2 regulates akt upon h2o2-dependent signaling activation in cancer cells. Cancers 2019, 11, 492. [Google Scholar] [CrossRef] [Green Version]
- Saxena, K.; Jolly, M.K. Acute vs. Chronic vs. cyclic hypoxia: Their differential dynamics, molecular mechanisms, and effects on tumor progression. Biomolecules 2019, 9, 339. [Google Scholar] [CrossRef] [Green Version]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-Inducible Factors and the Response to Hypoxic Stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, K.; Smith, K.; Gullick, W.J.; Harris, A.L. Mutant epidermal growth factor receptor enhances induction of vascular endothelial growth factor by hypoxia and insulin-like growth factor-1 via a Pl3 kinase dependent pathway. Br. J. Cancer 2001, 84, 1322–1329. [Google Scholar] [CrossRef] [Green Version]
- Laughner, E.; Taghavi, P.; Chiles, K.; Mahon, P.C.; Semenza, G.L. HER2 (neu) Signaling Increases the Rate of Hypoxia-Inducible Factor 1 (HIF-1) Synthesis: Novel Mechanism for HIF-1-Mediated Vascular Endothelial Growth Factor Expression. Mol. Cell. Biol. 2001, 21, 3995–4004. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.; Chiles, K.; Feldser, D.; Laughner, E.; Hanrahan, C.; Georgescu, M.M.; Simons, J.W.; Semenza, G.L. Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: Implications for tumor angiogenesis and therapeutics. Cancer Res. 2000, 60, 1541–1545. [Google Scholar]
- Labak, C.M.; Wang, P.Y.; Arora, R.; Guda, M.R.; Asuthkar, S.; Tsung, A.J.; Velpula, K.K. Glucose transport: Meeting the metabolic demands of cancer, and applications in glioblastoma treatment. Am. J. Cancer Res. 2016, 6, 1599–1608. [Google Scholar]
- Semenza, G.L. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanzey, M.; Abdul Rahim, S.A.; Oudin, A.; Dirkse, A.; Kaoma, T.; Vallar, L.; Herold-Mende, C.; Bjerkvig, R.; Golebiewska, A.; Niclou, S.P. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS ONE 2015, 10, e0123544. [Google Scholar] [CrossRef] [PubMed]
- Plate, K.H.; Breier, G.; Weich, H.A.; Risau, W. Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature 1992, 359, 845–848. [Google Scholar] [CrossRef]
- Plate, K.H.; Mennel, H.D. Vascular morphology and angiogenesis in glial tumors. Exp. Toxicol. Pathol. 1995, 47, 89–94. [Google Scholar] [CrossRef]
- Shweiki, D.; Itin, A.; Soffer, D.; Keshet, E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature 1992, 359, 843–845. [Google Scholar] [CrossRef]
- Friedman, H.S.; Prados, M.D.; Wen, P.Y.; Mikkelsen, T.; Schiff, D.; Abrey, L.E.; Yung, W.K.A.; Paleologos, N.; Nicholas, M.K.; Jensen, R.; et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J. Clin. Oncol. 2009, 27, 4733–4740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, S.; Nakagawa, K.; Harada, H.; Nagato, S.; Furukawa, K.; Teraoka, M.; Seno, T.; Oka, K.; Iwata, S.; Ohnishi, T. Silencing hypoxia-inducible factor-1α inhibits cell migration and invasion under hypoxic environment in malignant gliomas. Int. J. Oncol. 2007, 30, 793–802. [Google Scholar] [CrossRef] [Green Version]
- Wick, W.; Platten, M.; Weller, M. Glioma cell invasion: Regulation of metalloproteinase activity by TGF-beta. J. Neurooncol. 2001, 53, 177–185. [Google Scholar] [CrossRef]
- Madureira, P.A.; Hill, R.; Miller, V.A.; Wing, P.; Lee, K.; Waisman, D.M. Annexin A2 is a novel Cellular Redox Regulatory Protein involved in Tumorigenesis. Oncotarget 2011, 2, 1075–1093. [Google Scholar] [CrossRef]
- Madureira, P.A.; Waisman, D.M. Annexin A2: The importance of being redox sensitive. Int. J. Mol. Sci. 2013, 14, 3568–3594. [Google Scholar] [CrossRef] [Green Version]
- Kubala, M.H.; DeClerck, Y.A. The plasminogen activator inhibitor-1 paradox in cancer: A mechanistic understanding. Cancer Metastasis Rev. 2019, 38, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Osmak, M.; Vrhovec, I.; Skrk, J. Cisplatin resistant glioblastoma cells may have increased concentration of urokinase plasminogen activator and plasminogen activator inhibitor type 1. J. Neurooncol. 1999, 42, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Osmak, M.; Niksíc, D.; Brozović, A.; Ristov, A.A.; Vrhovec, I.; Skrk, J. Drug resistant tumor cells have increased levels of tumor markers for invasion and metastasis. Anticancer Res. 1999, 19, 3193–3197. [Google Scholar] [PubMed]
- Arai, Y.; Kubota, T.; Nakagawa, T.; Kabuto, M.; Sato, K.; Kobayashi, H. Production of urokinase-type plasminogen activator (u-PA) and plasminogen activator inhibitor-1 (PAI-1) in human brain tumours. Acta Neurochir. (Wien) 1998, 140, 377–386. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sawaya, R.; Mohanam, S.; Bindal, A.K.; Bruner, J.M.; Oka, K.; Rao, V.H.; Tomonaga, M.; Nicolson, G.L.; Rao, J.S. Expression and localization of urokinase-type plasminogen activator in human astrocytomas in vivo. Cancer Res. 1994, 54, 3656–3661. [Google Scholar]
- Zhang, X.; Bu, X.Y.; Zhen, H.N.; Fei, Z.; Zhang, J.N.; Fu, L.A. Expression and localisation of urokinase-type plasminogen activator gene in gliomas. J. Clin. Neurosci. 2000, 7, 116–119. [Google Scholar] [CrossRef]
- Alapati, K.; Kesanakurti, D.; Rao, J.S.; Dasari, V.R. uPAR and cathepsin B-mediated compartmentalization of JNK regulates the migration of glioma-initiating cells. Stem Cell Res. 2014, 12, 716–729. [Google Scholar] [CrossRef] [Green Version]
- Adachi, Y.; Chandrasekar, N.; Kin, Y.; Lakka, S.S.; Mohanam, S.; Yanamandra, N.; Mohan, P.M.; Fuller, G.N.; Fang, B.; Fueyo, J.; et al. Suppression of glioma invasion and growth by adenovirus-mediated delivery of a bicistronic construct containing antisense upar and sense p16 gene sequences. Oncogene 2002, 21, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Mori, T.; Abe, T.; Wakabayashi, Y.; Hikawa, T.; Matsuo, K.I.; Yamada, Y.; Kuwano, M.; Hori, S. Up-regulation of urokinase-type plasminogen activator and its receptor correlates with enhanced invasion activity of human glioma cells mediated by transforming growth factor-α or basic fibroblast growth factor. J. Neurooncol. 2000, 46, 115–123. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sawaya, R.; Mohanam, S.; Rao, V.H.; Bruner, J.M.; Nicolson, G.L.; Rao, J.S. Expression and localization of urokinase-type plasminogen activator receptor in human gliomas. Cancer Res. 1994, 54, 5016–5020. [Google Scholar]
- Krishnamachary, B.; Berg-Dixon, S.; Kelly, B.; Agani, F.; Feldser, D.; Ferreira, G.; Iyer, N.; LaRusch, J.; Pak, B.; Taghavi, P.; et al. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003, 63, 1138–1143. [Google Scholar] [PubMed]
- Kitamuro, T.; Takahashi, K.; Nakayama, M.; Murakami, O.; Hida, W.; Shirato, K.; Shibahara, S. Induction of adrenomedullin during hypoxia in cultured human glioblastoma cells. J. Neurochem. 2000, 75, 1826–1833. [Google Scholar] [CrossRef]
- Metellus, P.; Voutsinos-Porche, B.; Nanni-Metellus, I.; Colin, C.; Fina, F.; Berenguer, C.; Dussault, N.; Boudouresque, F.; Loundou, A.; Intagliata, D.; et al. Adrenomedullin expression and regulation in human glioblastoma, cultured human glioblastoma cell lines and pilocytic astrocytoma. Eur. J. Cancer 2011, 47, 1727–1735. [Google Scholar] [CrossRef] [PubMed]
- Heaton, J.; Lin, B.; Chang, J.K.; Steinberg, S.; Hyman, A.; Lippton, H. Pulmonary vasodilation to adrenomedullin: A novel peptide in humans. Am. J. Physiol. Hear. Circ. Physiol. 1995, 268. [Google Scholar] [CrossRef] [PubMed]
- Nikitenko, L.L.; MacKenzie, I.Z.; Rees, M.C.; Bicknell, R. Adrenomedullin is an autocrine regulator of endothelial growth in human endometrium. Mol. Hum. Reprod. 2000, 6, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Jougasaki, M.; Burnett, J.C. Adrenomedullin: Potential in physiology and pathophysiology. Life Sci. 2000, 66, 855–872. [Google Scholar] [CrossRef]
- Zhu, P.; Goh, Y.Y.; Chin, H.F.A.; Kersten, S.; Tan, N.S. Angiopoietin-like 4: A decade of research. Biosci. Rep. 2012, 32, 211–219. [Google Scholar] [CrossRef]
- Kolb, R.; Kluz, P.; Tan, Z.W.; Borcherding, N.; Bormann, N.; Vishwakarma, A.; Balcziak, L.; Zhu, P.; Davies, B.S.; Gourronc, F.; et al. Obesity-associated inflammation promotes angiogenesis and breast cancer via angiopoietin-like 4. Oncogene 2019, 38, 2351–2363. [Google Scholar] [CrossRef]
- Hu, K.; Babapoor-Farrokhran, S.; Rodrigues, M.; Deshpande, M.; Puchner, B.; Kashiwabuchi, F.; Hassan, S.J.; Asnaghi, L.; Handa, J.T.; Merbs, S.; et al. Hypoxia-inducible factor 1 upregulation of both VEGF and ANGPTL4 is required to promote the angiogenic phenotype in uveal melanoma. Oncotarget 2016, 7, 7816–7828. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Li, S.-Y.; Ji, F.-Y.; Zhao, Y.-F.; Zhong, Y.; Lv, X.-J.; Wu, X.-L.; Qian, G.-S. Role of Angptl4 in vascular permeability and inflammation. Inflamm. Res. 2014, 63, 13–22. [Google Scholar] [CrossRef]
- Tsai, Y.-T.; Wu, A.-C.; Yang, W.-B.; Kao, T.-J.; Chuang, J.-Y.; Chang, W.-C.; Hsu, T.-I. ANGPTL4 Induces TMZ Resistance of Glioblastoma by Promoting Cancer Stemness Enrichment via the EGFR/AKT/4E-BP1 Cascade. Int. J. Mol. Sci. 2019, 20, 5625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katanasaka, Y.; Kodera, Y.; Kitamura, Y.; Morimoto, T.; Tamura, T.; Koizumi, F. Epidermal growth factor receptor variant type III markedly accelerates angiogenesis and tumor growth via inducing c-myc mediated angiopoietin-like 4 expression in malignant glioma. Mol. Cancer 2013, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, T.R.; Eisenstat, D.D.; Gibson, S.B. BNIP3 (Bcl-2 19 kDa interacting protein) acts as transcriptional repressor of apoptosis-inducing factor expression preventing cell death in human malignant gliomas. J. Neurosci. 2009, 29, 4189–4199. [Google Scholar] [CrossRef] [PubMed]
- Burton, T.R.; Henson, E.S.; Azad, M.B.; Brown, M.; Eisenstat, D.D.; Gibson, S.B. BNIP3 acts as transcriptional repressor of death receptor-5 expression and prevents TRAIL-induced cell death in gliomas. Cell Death Dis. 2013, 4, e587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.L.; DeLay, M.; Jahangiri, A.; Molinaro, A.M.; Rose, S.D.; Carbonell, W.S.; Aghi, M.K. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012, 72, 1773–1783. [Google Scholar] [CrossRef] [Green Version]
- Mongiardi, M.P.; Savino, M.; Falchetti, M.L.; Illi, B.; Bozzo, F.; Valle, C.; Helmer-Citterich, M.; Ferrè, F.; Nasi, S.; Levi, A. c-MYC inhibition impairs hypoxia response in glioblastoma multiforme. Oncotarget 2016, 7, 33257–33271. [Google Scholar] [CrossRef]
- DeYoung, M.P.; Horak, P.; Sofer, A.; Sgroi, D.; Ellisen, L.W. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008, 22, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Laplante, M.; Sabatini, D.M. MTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [Green Version]
- Jawhari, S.; Ratinaud, M.-H.; Verdier, M. Glioblastoma, hypoxia and autophagy: A survival-prone “ménage-à-trois”. Cell Death Dis. 2016, 7, e2434. [Google Scholar] [CrossRef]
- Foltyn, M.; Luger, A.-L.; Lorenz, N.I.; Sauer, B.; Mittelbronn, M.; Harter, P.N.; Steinbach, J.P.; Ronellenfitsch, M.W. The physiological mTOR complex 1 inhibitor DDIT4 mediates therapy resistance in glioblastoma. Br. J. Cancer 2019, 120, 481–487. [Google Scholar] [CrossRef] [Green Version]
- Pinto, J.A.; Rolfo, C.; Raez, L.E.; Prado, A.; Araujo, J.M.; Bravo, L.; Fajardo, W.; Morante, Z.D.; Aguilar, A.; Neciosup, S.P.; et al. In silico evaluation of DNA Damage Inducible Transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Said, H.M.; Stein, S.; Hagemann, C.; Polat, B.; Staab, A.; Anacker, J.; Schoemig, B.; Theobald, M.; Flentje, M.; Vordermark, D. Oxygen-dependent regulation of NDRG1 in human glioblastoma cells in vitro and in vivo. Oncol. Rep. 2009, 21, 237–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Said, H.M.; Safari, R.; Al-Kafaji, G.; Ernestus, R.I.; Löhr, M.; Katzer, A.; Flentje, M.; Hagemann, C. Time- and oxygen-dependent expression and regulation of NDRG1 in human brain cancer cells. Oncol. Rep. 2017, 37, 3625–3634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonkeren, S.L.; Massen, M.; van der Horst, R.; Koch, A.; Vaes, N.; Melotte, V. Nervous NDRGs: The N-myc downstream–regulated gene family in the central and peripheral nervous system. Neurogenetics 2019, 20, 173–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiler, M.; Blaes, J.; Pusch, S.; Sahm, F.; Czabanka, M.; Luger, S.; Bunse, L.; Solecki, G.; Eichwald, V.; Jugold, M.; et al. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc. Natl. Acad. Sci. USA 2014, 111, 409–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Said Egr-1 is not upregulated in response to hypoxic and oxygenation conditions in human glioblastoma in vitro. Mol. Med. Rep. 2009, 2, 757–763. [CrossRef] [Green Version]
- Rong, Y.; Hu, F.; Huang, R.; Mackman, N.; Horowitz, J.M.; Jensen, R.L.; Durden, D.L.; Van Meir, E.G.; Brat, D.J. Early growth response gene-1 regulates hypoxia-induced expression of tissue factor in glioblastoma multiforme through hypoxia-inducible factor-1-independent mechanisms. Cancer Res. 2006, 66, 7067–7074. [Google Scholar] [CrossRef] [Green Version]
- Calogero, A.; Lombari, V.; De Gregorio, G.; Porcellini, A.; Ucci, S.; Arcella, A.; Caruso, R.; Gagliardi, F.M.; Gulino, A.; Lanzetta, G.; et al. Inhibition of cell growth by EGR-1 in human primary cultures from malignant glioma. Cancer Cell Int. 2004, 4. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Yao, J.; Mercola, D.; Adamson, E. The transcription factor EGR-1 directly transactivates the fibronectin gene and enhances attachment of human glioblastoma cell line U251. J. Biol. Chem. 2000, 275, 20315–20323. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Liu, Y.C.; Zheng, Y.Y.; Xu, J.; Zhang, Y.; Liu, W.L.; Li, Z.Y.; Huang, G.D.; Li, W.P. Furanodienone overcomes temozolomide resistance in glioblastoma through the downregulation of CSPG4-Akt-ERK signalling by inhibiting EGR1-dependent transcription. Phyther. Res. 2019, 33, 1736–1747. [Google Scholar] [CrossRef]
- Kim, J.W.; Kim, J.Y.; Kim, J.E.; Kim, S.K.; Chung, H.T.; Park, C.K. HOXA10 is associated with temozolomide resistance through regulation of the homologous recombinant DNA repair pathway in glioblastoma cell lines. Genes Cancer 2014, 5, 165–174. [Google Scholar] [CrossRef]
- Riddick, G.; Kotliarova, S.; Rodriguez, V.; Kim, H.S.; Linkous, A.; Storaska, A.J.; Ahn, S.; Walling, J.; Belova, G.; Fine, H.A. A Core Regulatory Circuit in Glioblastoma Stem Cells Links MAPK Activation to a Transcriptional Program of Neural Stem Cell Identity. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakakini, N.; Turchi, L.; Bergon, A.; Holota, H.; Rekima, S.; Lopez, F.; Paquis, P.; Almairac, F.; Fontaine, D.; Baeza-Kallee, N.; et al. A positive feed-forward loop associating EGR1 and PDGFA promotes proliferation and self-renewal in glioblastoma stem cells. J. Biol. Chem. 2016, 291, 10684–10699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Li, X.; Dong, D.; Zhang, B.; Xue, Y.; Shang, P. Transferrin receptor 1 in cancer: A new sight for cancer therapy. Am. J. Cancer Res. 2018, 8, 916–931. [Google Scholar] [PubMed]
- Yu, X.; Feng, L.; Liu, D.; Zhang, L.; Wu, B.; Jiang, W.; Han, Z.; Cheng, S. Quantitative proteomics reveals the novel co-expression signatures in early brain development for prognosis of glioblastoma multiforme. Oncotarget 2016, 7, 14161–14171. [Google Scholar] [CrossRef]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chédeville, A.L.; Lourdusamy, A.; Monteiro, A.R.; Hill, R.; Madureira, P.A. Investigating Glioblastoma Response to Hypoxia. Biomedicines 2020, 8, 310. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8090310
Chédeville AL, Lourdusamy A, Monteiro AR, Hill R, Madureira PA. Investigating Glioblastoma Response to Hypoxia. Biomedicines. 2020; 8(9):310. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8090310
Chicago/Turabian StyleChédeville, Agathe L., Anbarasu Lourdusamy, Ana Rita Monteiro, Richard Hill, and Patricia A. Madureira. 2020. "Investigating Glioblastoma Response to Hypoxia" Biomedicines 8, no. 9: 310. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines8090310