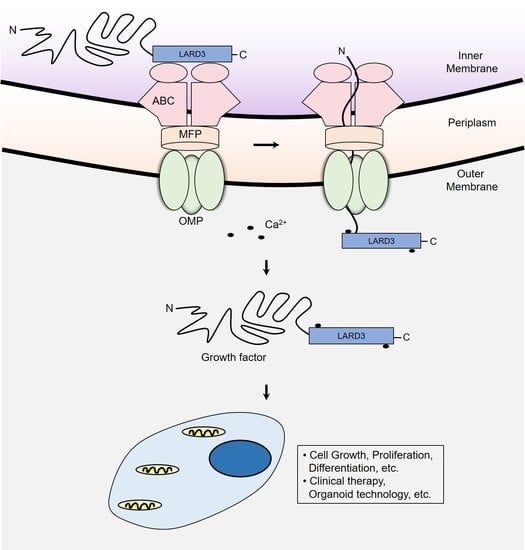
Utilizing the ABC Transporter for Growth Factor Production by fleQ Deletion Mutant of Pseudomonas fluorescens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Plasmid Construction
2.3. Identification of FliC Using Mass Spectrometry
2.4. P. fluorescens ΔfleQ Construction and Verification
2.5. Supercharging the Growth Factors
2.6. Recombinant Protein Expression and Secretion Analyses
2.7. Examining Recombinant Protein Secretion in Flask Culture
3. Results
3.1. Construction and Verification of P. fluorescens ΔfleQ Mutant
3.2. Comparison between Recombinant Protein Secretion of P. fluorescens Δtp and ΔfleQ
3.3. Expression and Secretion of Recombinant Growth Factors in P. fluorescens ΔfleQ
3.4. Flask-Volume Preparation of P. fluorescens ΔfleQ
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Son, M.Y.; Oh, M.J.; Han, S.B.; Park, K.H.; Kim, J.G.; Ahn, J.H. Lipase and protease double-deletion mutant of Pseudomonas fluorescens suitable for extracellular protein production. Appl. Environ. Microbiol. 2012, 78, 8454–8462. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.H.; Pan, J.G.; Rhee, J.S. Identification of the tliDEF ABC transporter specific for lipase in Pseudomonas fluorescens SIK W1. J. Bacteriol. 1999, 181, 1847–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sheikh, M.A.; El-Kazzaz, S.A.; Hafez, E.E.; Madkour, S.A.; El-Gayyar, S.M. Detection purification, and identification of siderophores produced by Pseudomonas fluorescens isolates using SDS-page and HPLC. J. Plant Prot. Res. 2011, 2, 691–705. [Google Scholar] [CrossRef]
- Haas, D.; Keel, C. Regulation of Antibiotic Production in Root-Colonizing Pseudomonas spp. and Relevance for Biological Control of Plant Disease. Ann. Rev. Phytopathol. 2003, 41, 117–153. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.R.B.; Dos Santos, V.A.P.M.; Pieper, D.H.; Ramos, J.L.; Palleron, N.J. Prokaryotes: A Handbook on the Biology of Bacteria, 3rd ed.; Springer: New York, NY, USA, 2006; Volume 6. [Google Scholar]
- Raaijmakers, J.M.; Vlami, M.; de Souza, J.T. Antibiotic production by bacterial biocontrol agents. Antonie Leeuwenhoek 2002, 81, 537. [Google Scholar] [CrossRef]
- Malik, D.K.; Sindhu, S.S. Production of indole acetic acid by Pseudomonas sp.: Effect of coinoculation with Mesorhizobium sp. Cicer on nodulation and plant growth of chickpea (Cicer arietinum). Physiol. Mol. Biol. Plants 2011, 17, 25–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.; Moon, Y.; Ryoo, J.; Kim, N.; Cho, H.; Ahn, J.H. Identification of the minimal region in lipase ABC transporter recognition domain of Pseudomonas fluorescens for secretion and fluorescence of green fluorescent protein. Microb. Cell Fact. 2012, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- Moens, S.; Vanderleyden, J. Functions of Bacterial Flagella. Crit. Rev. Microbiol. 1996, 22, 67–100. [Google Scholar] [CrossRef]
- Dasgupta, N.; Wolfgang, M.C.; Goodman, A.L.; Arora, S.K.; Jyot, J.; Lory, S.; Ramphal, R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 809–824. [Google Scholar] [CrossRef] [Green Version]
- Arora, S.K.; Ritchings, B.W.; Almira, E.C.; Lory, S.; Ramphal, R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1997, 179, 5574–5581. [Google Scholar] [CrossRef] [Green Version]
- Kieboom, J.; Bruinenberg, R.; Keizer-Gunnink, I.; de Bont, J.A. Transposon mutations in the flagella biosynthetic pathway of the solvent-tolerant Pseudomonas putida S12 result in a decreased expression of solvent efflux genes. FEMS Microbiol. Lett. 2001, 198, 117–122. [Google Scholar] [CrossRef]
- Jiménez-Fernández, A.; López-Sánchez, A.; Jiménez-Díaz, L.; Navarrete, B.; Calero, P.; Platero, A.I.; Govantes, F. Complex Interplay between FleQ, Cyclic Diguanylate and Multiple σ Factors Coordinately Regulates Flagellar Motility and Biofilm Development in Pseudomonas putida. PLoS ONE 2016, 11, e0163142. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Hu, X.; Wang, J.; Li, H.; Wang, X. Impact of mycolic acid deficiency on cells of Corynebacterium glutamicum ATCC13869. Biotechnol. Appl. Biochem. 2018, 65, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Robleto, E.A.; López-Hernández, I.; Silby, M.W.; Levy, S.B. Genetic analysis of the AdnA regulon in Pseudomonas fluorescens: Nonessential role of flagella in adhesion to sand and biofilm formation. J. Bacteriol. 2003, 185, 453–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capdevila, S.; Martínez-Granero, F.M.; Sánchez-Contreras, M.; Rivilla, R.; Martín, M. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 2004, 150, 3889–3897. [Google Scholar] [CrossRef] [Green Version]
- Redondo-Nieto, M.; Lloret, J.; Larenas, J.; Barahona, E.; Navazo, A.; Martínez-Granero, F.; Capdevila, S.; Rivilla, R.; Martín, M. Transcriptional organization of the region encoding the synthesis of the flagellar filament in Pseudomonas fluorescens. J. Bacteriol. 2008, 190, 4106–4109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jyot, J.; Dasgupta, N.; Ramphal, R. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 2002, 184, 5251–5260. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, A.C.; Briquez, P.S.; Hubbell, J.A.; Cochran, J.R. Engineering growth factors for regenerative medicine applications. Acta Biomater. 2016, 30, 1–12. [Google Scholar] [CrossRef]
- Fitzpatrick, R.E. Endogenous Growth Factors as Cosmeceuticals. Dermatol. Surg. 2005, 31, 827–831. [Google Scholar] [CrossRef]
- Schuldiner, M.; Yanuka, O.; Itskovitz-Eldor, J.; Melton, D.A.; Benvenisty, N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2000, 97, 11307. [Google Scholar] [CrossRef] [Green Version]
- Byun, H.; Park, J.; Kim, S.C.; Ahn, J.H. A lower isoelectric point increases signal sequence-mediated secretion of recombinant proteins through a bacterial ABC transporter. J. Biol. Chem. 2017, 292, 19782–19791. [Google Scholar] [CrossRef] [Green Version]
- Pappas, G.; Karavasilis, V.; Christou, L.; Tsianos, E.V. Pseudomonas fluorescens infections in clinical practice. Scand. J. Infect. Dis. 2006, 38, 68–70. [Google Scholar] [CrossRef]
- Ryu, J.; Lee, U.; Park, J.; Yoo, D.H.; Ahn, J.H. A vector system for ABC transporter-mediated secretion and purification of recombinant proteins in Pseudomonas species. Appl. Environ. Microbiol. 2015, 81, 1744–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Xiao, Y.; Nie, H.; Huang, Q.; Chen, W. Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol. Res. 2017, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Phillips, K.J.; Liu, D.R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 2007, 129, 10110–10112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Der, B.S.; Kluwe, C.; Miklos, A.E.; Jacak, R.; Lyskov, S.; Gray, J.J.; Georgiou, G.; Ellington, A.D.; Kuhlman, B. Alternative Computational Protocols for Supercharging Protein Surfaces for Reversible Unfolding and Retention of Stability. PLoS ONE 2013, 8, e64363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Ryu, J.; Byun, H.; Park, J.P.; Park, J.; Noh, K.H.; Chung, J.H.; Lee, H.; Ahn, J.H. Tat-Dependent Heterologous Secretion of Recombinant Tyrosinase by Pseudomonas fluorescens Is Aided by a Translationally Fused Caddie Protein. Appl. Environ. Microbiol. 2019, 85, e01350-19. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Goker, M.; Martin, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolaidis, M.; Mossialos, D.; Oliver, S.G.; Amoutzias, G.D. Comparative Analysis of the Core Proteomes among the Pseudomonas Major Evolutionary Groups Reveals Species-Specific Adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis. Diversity 2020, 12, 289. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Arrebola, E.; Martínez-Granero, F.; García-Méndez, S.; Muriel, C.; Blanco-Romero, E.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Classification of Isolates from the Pseudomonas fluorescens Complex into Phylogenomic Groups Based in Group-Specific Markers. Front. Microbiol. 2017, 8, 413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagorn, C.A.; Mijouin, L.; Hillion, M.; Duclairoir-Poc, C.; Chevalier, S.; Taupin, L.; Orange, N.; Feuilloley, M.G. Effect of GABA, a bacterial metabolite, on Pseudomonas fluorescens surface properties and cytotoxicity. Int. J. Mol. Sci. 2013, 14, 12186–12204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EPA. Guidelines for Exposure Assessment; U.S. Environmental Protection Agency: Washington, DC, USA, 1992.
- Anderson, J.A.; Staley, J.; Challender, M.; Heuton, J. Safety of Pseudomonas chlororaphis as a gene source for genetically modified crops. Transgenic Res. 2018, 27, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, J.F. Guide to Plant Pathogenic Bacteria; CAB International Farnham Royal: Slough, UK, 1986; p. 332. [Google Scholar]
- Squires, C.H.; Retallack, D.; Chew, L.; Ramseier, T.; Schneider, J.C. Heterologous Protein Production in P. fluorescens. BioProcess Int. 2004, 2, e58. [Google Scholar]
- Retallack, D.; Schneider, J.C.; Chew, L.; Ramseier, T.; Allen, J.; Patkar, A.; Squires, C.; Talbot, H.; Mitchell, J. Pseudomonas fluorescens—A robust expression platform for pharmaceutical protein production. Microb. Cell Fact. 2006, 5, S28. [Google Scholar] [CrossRef]
- Jin, H.; Cantin, G.T.; Maki, S.; Chew, L.C.; Resnick, S.M.; Ngai, J.; Retallack, D.M. Soluble periplasmic production of human granulocyte colony-stimulating factor (G-CSF) in Pseudomonas fluorescens. Protein Expr. Purif. 2011, 78, 69–77. [Google Scholar] [CrossRef]
- Ma, Q.; Zhai, Y.; Schneider, J.C.; Ramseier, T.M.; Saier, M.H., Jr. Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens. Biochim. Biophys. Acta 2003, 1611, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Cascales, E. The type VI secretion toolkit. EMBO Rep. 2008, 9, 735–741. [Google Scholar] [CrossRef] [Green Version]
- Kostakioti, M.; Newman, C.L.; Thanassi, D.G.; Stathopoulos, C. Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 2005, 187, 4306–4314. [Google Scholar] [CrossRef] [Green Version]
- Jasinski, M.; Ducos, E.; Martinoia, E.; Boutry, M. The ATP-Binding Cassette Transporters: Structure, Function, and Gene Family Comparison between Rice and Arabidopsis. Plant Physiol. 2003, 131, 1169. [Google Scholar] [CrossRef] [Green Version]
- Biemans-Oldehinkel, E.; Doeven, M.K.; Poolman, B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2006, 580, 1023–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, I.R.; Navarro-Garcia, F.; Desvaux, M.; Fernandez, R.C.; Ala Aldeen, D. Type V Protein Secretion Pathway: The Autotransporter Story. Microbiol. Mol. Biol. Rev. 2004, 68, 692. [Google Scholar] [CrossRef] [Green Version]
- Wrigley, S.; Arafa, D.; Tropea, D. Insulin-Like Growth Factor 1: At the Crossroads of Brain Development and Aging. Front. Cell. Neurosci. 2017, 11, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre, G.A.; De Ita, J.R.; de la Garza, R.G.; Castilla-Cortazar, I. Insulin-like growth factor-1 deficiency and metabolic syndrome. J. Transl. Med. 2016, 14, 3. [Google Scholar] [CrossRef] [Green Version]
- O’Dell, S.D.; Day, I.N. Insulin-like growth factor II (IGF-II). Int. J. Biochem. Cell Biol. 1998, 30, 767–771. [Google Scholar] [CrossRef]
- Rodríguez, S.; Gaunt, T.R.; O’Dell, S.D.; Chen, X.-H.; Gu, D.; Hawe, E.; Miller, G.J.; Humphries, S.E.; Day, I.N.M. Haplotypic analyses of the IGF2-INS-TH gene cluster in relation to cardiovascular risk traits. Hum. Mol. Genet. 2004, 13, 715–725. [Google Scholar] [CrossRef] [Green Version]
- Faienza, M.F.; Santoro, N.; Lauciello, R.; Calabrò, R.; Giordani, L.; Di Salvo, G.; Ventura, A.; Delvecchio, M.; Perrone, L.; Del Giudice, E.M.; et al. IGF2 Gene Variants and Risk of Hypertension in Obese Children and Adolescents. Pediatr. Res. 2010, 67, 340–344. [Google Scholar] [CrossRef] [Green Version]
- Levi-Montalcini, R. The nerve growth factor 35 years later. Science 1987, 237, 1154–1162. [Google Scholar] [CrossRef]
- Aloe, L.; Rocco, M.L.; Balzamino, B.O.; Micera, A. Nerve Growth Factor: A Focus on Neuroscience and Therapy. Curr. Neuropharmacol. 2015, 13, 294–303. [Google Scholar] [CrossRef] [Green Version]
- Fahnestock, M.; Yu, G.; Michalski, B.; Mathew, S.; Colquhoun, A.; Ross, G.M.; Coughlin, M.D. The nerve growth factor precursor proNGF exhibits neurotrophic activity but is less active than mature nerve growth factor. J. Neurochem. 2004, 89, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Fahnestock, M.; Yu, G.; Coughlin, M.D. ProNGF: A neurotrophic or an apoptotic molecule? Prog. Brain Res. 2004, 146, 101–110. [Google Scholar] [PubMed]
- Banai, S.; Jaklitsch, M.T.; Casscells, W.; Shou, M.; Shrivastav, S.; Correa, R.; Epstein, S.E.; Unger, E.F. Effects of acidic fibroblast growth factor on normal and ischemic myocardium. Circ. Res. 1991, 69, 76–85. [Google Scholar] [CrossRef] [Green Version]
- Beenken, A.; Mohammadi, M. The FGF family: Biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009, 8, 235–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, L.; Ding, L.; Lan, J.; Niu, J.; He, Y.; Song, L. Fibroblast Growth Factor-1 Improves Insulin Resistance via Repression of JNK-Mediated Inflammation. Front. Pharmacol. 2019, 10, 1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Zheng, L.; Yuan, Q.; Zhen, G.; Crane, J.L.; Zhou, X.; Cao, X. Transforming growth factor-β in stem cells and tissue homeostasis. Bone Res. 2018, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Flanders, K.C.; Burmester, J.K. Medical applications of transforming growth factor-beta. Clin. Med. Res. 2003, 1, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Vassalli, P. The pathophysiology of tumor necrosis factors. Annu. Rev. Immunol. 1992, 10, 411–452. [Google Scholar] [CrossRef] [PubMed]
- Laddha, N.C.; Dwivedi, M.; Gani, A.R.; Mansuri, M.S.; Begum, R. Tumor necrosis factor B (TNFB) genetic variants and its increased expression are associated with vitiligo susceptibility. PLoS ONE 2013, 8, e81736. [Google Scholar] [CrossRef] [Green Version]
- Beutler, B.; Bazzoni, F. TNF, apoptosis and autoimmunity: A common thread? Blood Cells Mol. Dis. 1998, 24, 216–230. [Google Scholar] [CrossRef] [PubMed]





| Primer ID | Primer Sequence for PCR |
|---|---|
| EcoRI_ΔfleQ_F1f | F 5′ GGG GAATTC AGAGCCTGCTCGCGGTTTA 3′ |
| ΔfleQ_F1r | R 5′ TAGTCCTTGA ACGGCTCGATGACAATGAGC 3′ |
| ΔfleQ_F2f | F 5′ ATCGAGCCGT TCAAGGACTACCTCGGCAAC 3′ |
| ΔfleQ_F2r_HindIII | R 5′ GGG AAGCTT TCGGAGACCCGGGCTTCC 3′ |
| F-PCR | F 5′ GCCGACATGATCGACGAAG 3′ R 5′ TCCAGGGAACGGGTGGCG 3′ |
| Recombinant Protein | Mw (kDa) * | Concentration (mg/L) |
|---|---|---|
| IGFI(−) | 20.6 | 5.6 |
| IGFII(−) | 20.2 | 6.0 |
| βNGF(−) | 25.6 | 8.6 |
| FGF1(−) | 28.1 | 6.6 |
| TGFβ(−) | 25.3 | 1.1 |
| TNFβ(−) | 30.7 | 0.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabia, B.-U.; Bingwa, J.; Park, J.; Hieu, N.-M.; Ahn, J.-H. Utilizing the ABC Transporter for Growth Factor Production by fleQ Deletion Mutant of Pseudomonas fluorescens. Biomedicines 2021, 9, 679. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9060679
Fabia B-U, Bingwa J, Park J, Hieu N-M, Ahn J-H. Utilizing the ABC Transporter for Growth Factor Production by fleQ Deletion Mutant of Pseudomonas fluorescens. Biomedicines. 2021; 9(6):679. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9060679
Chicago/Turabian StyleFabia, Benedict-Uy, Joshua Bingwa, Jiyeon Park, Nguyen-Mihn Hieu, and Jung-Hoon Ahn. 2021. "Utilizing the ABC Transporter for Growth Factor Production by fleQ Deletion Mutant of Pseudomonas fluorescens" Biomedicines 9, no. 6: 679. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9060679