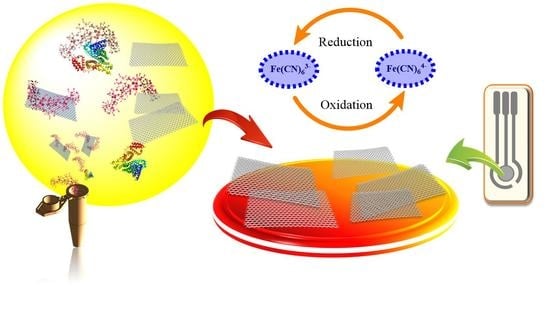
Immobilization-Free Electrochemical Sensor Coupled with a Graphene-Oxide-Based Aptasensor for Glycated Albumin Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Preparation of Electrodes
2.3. Preparation of the GO-Aptasensor Complex
2.4. Electrochemical Measurement
2.5. Fluorescence Measurement
3. Results and Discussion
3.1. Electrochemical Characterization
3.2. Fluorescence Characterization of the Developed Aptasensor Components
3.3. Performance of the Developed Electrochemical Aptasensor
3.4. Specificity, Reproducibility, and Stability of the Developed Electrochemical Aptasensor
3.5. Application of the Developed Electrochemical Aptasensor for the Detection of GHSA in Clinical Samples
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaspers, L.; Colpani, V.; Chaker, L.; van der Lee, S.J.; Muka, T.; Imo, D.; Mendis, S.; Chowdhury, R.; Bramer, W.M.; Falla, A.; et al. The global impact of non-communicable diseases on households and impoverishment: A systematic review. Eur. J. Epidemiol. 2014, 30, 163–188. [Google Scholar] [CrossRef]
- Cho, N.H.; Shaw, J.E.; Karuranga, S.; Huang, Y.; da Rocha Fernandes, J.D.; Ohlrogge, A.W.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018, 138, 271–281. [Google Scholar] [CrossRef]
- Wagner, K.H.; Brath, H. A global view on the development of noncommunicable diseases. Prev. Med. 2012, 54, S38–S41. [Google Scholar] [CrossRef]
- Van Dieren, S.; Beulens, J.W.J.; Van Der Schouw, Y.T.; Grobbee, D.E.; Neal, B. The global burden of diabetes and its complications: An emerging pandemic. Eur. J. Cardiovasc. Prev. Rehabil. 2010, 17, s3–s8. [Google Scholar]
- Piero, M.N. Diabetes mellitus-A devastating metabolic disorder. Asian J. Biomed. Pharm. Sci. 2015, 4, 1–7. [Google Scholar] [CrossRef]
- Stewart, B.W.; Wild, C.P. The global economic burden of non-communicable diseases. World Cancer Rep. 2014, 2014, 1–8. [Google Scholar]
- Boutayeb, A.; Boutayeb, S. The burden of noncommunicable diseases in developing countries. Int. J. Equity Health. 2005, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gan, T.; Liu, X.; Xu, G. Glycated albumin versus HbA1c in the evaluation of glycemic control in patients with diabetes and CKD. Kidney Int. Rep. 2018, 3, 542–554. [Google Scholar] [CrossRef] [Green Version]
- Alqahtani, N.; Khan, W.A.G.; Alhumaidi, M.H.; Ahmed, Y.A.A.R. Use of glycated hemoglobin in the diagnosis of diabetes mellitus and pre-diabetes and role of fasting plasma glucose, oral glucose tolerance test. Int. J. Prev. Med. 2013, 4, 1025–1029. [Google Scholar]
- Kosecki, S.M.; Rodgers, P.T.; Adams, M.B. Glycemic monitoring in diabetics with sickle cell plus β-thalassemia hemoglobinopathy. Ann. Pharmacother. 2005, 39, 1557–1560. [Google Scholar] [CrossRef]
- Inaba, M.; Okuno, S.; Kumeda, Y.; Yamada, S.; Imanishi, Y.; Tabata, T.; Okamura, M.; Okada, S.; Yamakawa, T.; Ishimura, E.; et al. Glycated albumin is a better glycemic indicator than glycated hemoglobin values in hemodialysis patients with diabetes: Effect of anemia and erythropoietin injection. J. Am. Soc. Nephrol. 2007, 18, 896–903. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Datta, D.; Cheema, M.; Dutta, M.; Stroscio, M.A. Aptasensor based optical detection of glycated albumin for diabetes mellitus diagnosis. Nanotechnology 2017, 43, 1–16. [Google Scholar] [CrossRef]
- Ciaccio, M. Introduction of glycated albumin in clinical practice. J. Lab. Precis. Med. 2019, 4, 28. [Google Scholar] [CrossRef]
- Peacock, T.P.; Shihabi, Z.K.; Bleyer, A.J.; Dolbare, E.L.; Byers, J.R.; Knovich, M.A.; Calles-Escandon, J.; Russell, G.B.; Freedman, B.I. Comparison of glycated albumin and hemoglobin A1c levels in diabetic subjects on hemodialysis. Kidney Int. 2008, 73, 1062–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, K.; Sakamoto, Y.; Kawasaki, Y.; Miyake, T.; Tanaka, K.; Urata, T.; Katayama, Y.; Ueda, S.; Horiuchi, S. Determination of glycated albumin by enzyme-linked boronate immunoassay (ELBIA). Clin. Chem. 1998, 44, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Goto, A.; Kishimoto, M.; Tsujimoto, T.; Yamamoto-Honda, R.; Noto, H.; Kajio, H.; Terauchi, Y.; Noda, M. Possible discrepancy of HbA1c values and its assessment among patients with chronic renal failure, hemodialysis and other diseases. Clin. Exp. Nephrol. 2015, 19, 1179–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dingari, N.C.; Horowitz, G.L.; Kang, J.W.; Dasari, R.R.; Barman, I. Raman spectroscopy provides a powerful diagnostic tool for accurate determination of albumin glycation. PLoS ONE 2012, 7, e32406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinton, D.J.S.; Ames, J.M. Analysis of glycated protein by capillary electrophoresis. Int. Congr. Ser. 2002, 1245, 471–474. [Google Scholar] [CrossRef]
- Apiwat, C.; Luksirikul, P.; Kankla, P.; Pongprayoon, P.; Treerattrakoon, K.; Paiboonsukwong, K.; Fucharoen, S.; Dharakul, T.; Japrung, D. Graphene based aptasensor for glycated albumin in diabetes mellitus diagnosis and monitoring. Biosens. Bioelectron. 2016, 82, 140–145. [Google Scholar] [CrossRef]
- Bunyarataphan, S.; Dharakul, T.; Fucharoen, S.; Paiboonsukwong, K.; Japrung, D. Glycated albumin measurement using an eletrochemical aptasensor for screening and monitoring of diabetes mellitus. Electroanalysis 2019, 31, 2254–2261. [Google Scholar] [CrossRef]
- Rao, C.N.R.; Sood, A.K.; Subrahmanyam, K.S.; Govindaraj, A. Graphene: The new two-dimensional nanomaterial. Angew Chemie Int. Ed. 2009, 48, 7752–7777. [Google Scholar] [CrossRef] [PubMed]
- Loo, A.H.; Bonanni, A.; Pumera, M. An insight into the hybridization mechanism of hairpin DNA physically immobilized on chemically modified graphenes. Analyst 2013, 138, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Loo, A.H.; Bonanni, A.; Pumera, M. Inherently electroactive graphene oxide nanoplatelets as labels for specific protein-target recognition. Nanoscale 2013, 5, 7844–7848. [Google Scholar] [CrossRef] [PubMed]
- Chawjiraphan, W.; Apiwat, C.; Segkhoonthod, K.; Treerattrakoon, K.; Pinpradup, P.; Sathirapongsasuti, N.; Pongprayoon, P.; Luksirikul, P.; Isarankura-Na-Ayudhya, P.; Japrung, D. Sensitive detection of albuminuria by graphene oxide-mediated fluorescence quenching aptasensor. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 231, 118–128. [Google Scholar] [CrossRef]
- Chawjiraphan, W.; Apiwat, C.; Segkhoonthod, K.; Treerattrakoon, K.; Pinpradup, P.; Sathirapongsasuti, N.; Pongprayoon, P.; Luksirikul, P.; Isarankura-Na-Ayudhya, P.; Japrung, D. Albuminuria detection using graphene oxide-mediated fluorescence quenching aptasensor. MethodsX 2020, 7, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Peña-Bahamonde, J.; Nguyen, H.N.; Fanourakis, S.K.; Rodrigues, D.F. Recent advances in graphene-based biosensor technology with applications in life sciences. J. Nanobiotechnol. 2018, 16, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Pan, G.; Avent, N.D.; Lowry, R.B.; Madgett, T.E.; Waines, P.L. Graphene electrode modified with electrochemically reduced graphene oxide for label-free DNA detection. Biosens. Bioelectron. 2015, 72, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Farzadfard, A.; Shayeh, J.S.; Habibi-Rezaei, M.; Omidi, M. Modification of reduced graphene/Au-aptamer to develop an electrochemical based aptasensor for measurement of glycated albumin. Talanta 2020, 211, 120722. [Google Scholar] [CrossRef]
- Inoue, Y.; Inoue, M.; Saito, M.; Yoshikawa, H.; Tamiya, E. Sensitive detection of glycated albumin in human serum albumin using electrochemiluminescence. Anal. Chem. 2017, 89, 5909–5915. [Google Scholar] [CrossRef] [PubMed]
- Panman, W.; Japrung, D.; Pongprayoon, P. Exploring the interactions of a DNA aptamer with human serum albumins: Simulation studies. J. Biomol. Struct. Dyn. 2016, 35, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Jaroenram, W.; Kampeera, J.; Arunrut, N.; Karuwan, C.; Sappat, A.; Khumwan, P.; Jaitrong, S.; Boonnak, K.; Prammananan, T.; Chaiprasert, A.; et al. Graphene-based electrochemical genosensor incorporated loop-mediated isothermal amplification for rapid on-site detection of Mycobacterium tuberculosis. J. Pharm. Biomed. Anal. 2020, 186, 113333. [Google Scholar] [CrossRef]








| Aptamer Name | Nucleotide Sequence |
|---|---|
| Modified G8-FAM | 5′-/56-FAM/-TGCGGTTCGTGCGGTTGTAGTAC-3′ |
| Modified G8 | 5′-TGCGGTTCGTGCGGTTGTAGTAC-3′ |
| Method Used | Detection Limit | Assay Time | Reference |
|---|---|---|---|
| Raman spectroscopy-based sensor | 13.7 µM | 1 h | [17] |
| Graphene-based optical aptasensor | 50 µg/mL | >30 min | [19] |
| Electrochemical aptasensor | 0.07 µg/mL | >24 h | [28] |
| Electrochemiluminescence | 6.6 µg/mL | >2 h | [29] |
| Optical aptasensor | 0.067 µg/mL | >2 h | [12] |
| Electrochemical aptasensor | 3 ng/mL | >17 h | [20] |
| Electrochemical GO-aptasensor | 8.70 ng/mL | 30 min | This study |
| Sample Source | GHSA Concentration (mg/mL) | |||
|---|---|---|---|---|
| Mean | SD | Minimum | Maximum | |
| Normal | 12.18 | 2.58 | 9.47 | 14.61 |
| Diabetics | 36.23 | 8.07 | 27.71 | 44.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waiwinya, W.; Putnin, T.; Pimalai, D.; Chawjiraphan, W.; Sathirapongsasuti, N.; Japrung, D. Immobilization-Free Electrochemical Sensor Coupled with a Graphene-Oxide-Based Aptasensor for Glycated Albumin Detection. Biosensors 2021, 11, 85. https://0-doi-org.brum.beds.ac.uk/10.3390/bios11030085
Waiwinya W, Putnin T, Pimalai D, Chawjiraphan W, Sathirapongsasuti N, Japrung D. Immobilization-Free Electrochemical Sensor Coupled with a Graphene-Oxide-Based Aptasensor for Glycated Albumin Detection. Biosensors. 2021; 11(3):85. https://0-doi-org.brum.beds.ac.uk/10.3390/bios11030085
Chicago/Turabian StyleWaiwinya, Wassa, Thitirat Putnin, Dechnarong Pimalai, Wireeya Chawjiraphan, Nuankanya Sathirapongsasuti, and Deanpen Japrung. 2021. "Immobilization-Free Electrochemical Sensor Coupled with a Graphene-Oxide-Based Aptasensor for Glycated Albumin Detection" Biosensors 11, no. 3: 85. https://0-doi-org.brum.beds.ac.uk/10.3390/bios11030085