Sequestration of 9-Hydroxystearic Acid in FAHFA (Fatty Acid Esters of Hydroxy Fatty Acids) as a Protective Mechanism for Colon Carcinoma Cells to Avoid Apoptotic Cell Death
Abstract
:1. Introduction
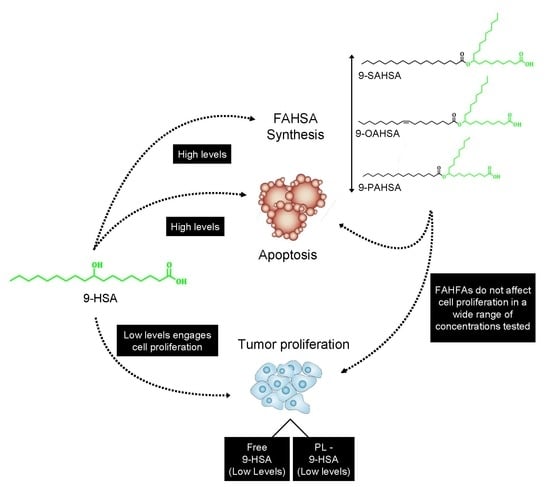
2. Results and Discussion
3. Materials and Methods
3.1. Reagents
3.2. Patients and Sampling Procedures
3.3. Cell Lines
3.4. Detection of Apoptosis
3.5. Immunoblot Analyses
3.6. Real-Time PCR
3.7. Gas Chromatography/Mass Spectrometry (GC/MS) Analyses
3.8. Liquid Chromatography/Mass Spectrometry (LC/MS) Analyses
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Araghi, M.; Soerjomataram, I.; Jenkins, M.; Brierley, J.; Morris, E.; Bray, F.; Arnold, M. Global trends in colorectal cancer mortality: Projections to the year 2035. Int. J. Cancer 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- De Visser, K.E.; Eichten, A.; Coussens, L.M. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer 2006, 6, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Mohme, M.; Riethdorf, S.; Pantel, K. Circulating and disseminated tumour cells—Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2016, 14, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2017, 18, 33–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, G.; Casali, E.; Spisni, A.; Masotti, L. Identification of the peroxidation product hydroxystearic acid in Lewis lung carcinoma cells. Biochem. Biophys. Res. Commun. 1991, 178, 1260–1265. [Google Scholar] [CrossRef]
- Masotti, L.; Casali, E.; Gesmundo, N. Influence of hydroxystearic acid on in vitro cell proliferation. Mol. Asp. Med. 1993, 14, 209–215. [Google Scholar] [CrossRef]
- Calonghi, N.; Cappadone, C.; Pagnotta, E.; Farruggia, G.; Buontempo, F.; Boga, C.; Brusa, G.L.; Santucci, M.A.; Masotti, L. 9-Hydroxystearic acid upregulates p21WAF1 in HT29 cancer cells. Biochem. Biophys. Res. Commun. 2004, 314, 138–142. [Google Scholar] [CrossRef]
- Calonghi, N.; Cappadone, C.; Pagnotta, E.; Boga, C.; Bertucci, C.; Fiori, J.; Tasco, G.; Casadio, R.; Masotti, L. Histone deacetylase 1: A target of 9-hydroxystearic acid in the inhibition of cell growth in human colon cancer. J. Lipid Res. 2005, 46, 1596–1603. [Google Scholar] [CrossRef]
- Calonghi, N.; Pagnotta, E.; Parolin, C.; Tognoli, C.; Boga, C.; Masotti, L. 9-Hydroxystearic acid interferes with EGF signalling in a human colon adenocarcinoma. Biochem. Biophys. Res. Commun. 2006, 342, 585–588. [Google Scholar] [CrossRef]
- Parolin, C.; Calonghi, N.; Presta, E.; Boga, C.; Caruana, P.; Naldi, M.; Andrisano, V.; Masotti, L.; Sartor, G. Mechanism and stereoselectivity of HDAC inhibition by (R)-9-hydroxystearic acid in colon cancer. Biochim. Biophys. Acta 2012, 1821, 1334–1340. [Google Scholar] [CrossRef] [PubMed]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef] [Green Version]
- Moraes-Vieira, P.M.; Saghatelian, A.; Kahn, B.B. GLUT4 expression in adipocytes regulates de novo lipogenesis and levels of a novel class of lipids with antidiabetic and anti-inflammatory effects. Diabetes 2016, 65, 1808–1815. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Thomas, C.P.; Morgan, L.T.; Maskrey, B.H.; Murphy, R.C.; Kühn, H.; Hazen, S.L.; Goodall, A.H.; Hamali, H.A.; Collins, P.W.; O’Donnell, V.B. Phospholipid-esterified eicosanoids are generated in agonist-activated human platelets and enhance tissue factor-dependent thrombin generation. J. Biol. Chem. 2010, 285, 6891–6903. [Google Scholar] [CrossRef]
- Slatter, D.A.; Aldrovandi, M.; O’Connor, A.; Allen, S.M.; Brasher, C.J.; Murphy, R.C.; Mecklemann, S.; Ravi, S.; Darley-Usmar, V.; O’Donnell, V.B. Mapping the human platelet lipidome reveals cytosolic phospholipase A2 as a regulator of mitochondrial bioenergetics during activation. Cell Metab. 2016, 23, 930–944. [Google Scholar] [CrossRef]
- O’Donnell, V.B.; Murphy, R.C. Directing eicosanoid esterification into phospholipids. J. Lipid Res. 2017, 58, 837–839. [Google Scholar] [CrossRef]
- Pulfer, M.; Murphy, R.C. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003, 22, 332–364. [Google Scholar] [CrossRef]
- Astudillo, A.M.; Balboa, M.A.; Balsinde, J. Selectivity of phospholipid hydrolysis by phospholipase A2 enzymes in activated cells leading to polyunsaturated fatty acid mobilization. Biochim. Biophys. Acta 2019, 1864, 772–783. [Google Scholar] [CrossRef]
- Casas, J.; Gijón, M.A.; Vigo, A.G.; Crespo, M.S.; Balsinde, J.; Balboa, M.A. Phosphatidylinositol 4, 5-bisphosphate anchors cytosolic group IVA phospholipase A2 to perinuclear membranes and decreases its calcium requirement for translocation in live cells. Mol. Biol. Cell 2006, 17, 155–162. [Google Scholar] [CrossRef]
- Guijas, C.; Rodríguez, J.P.; Rubio, J.M.; Balboa, M.A.; Balsinde, J. Phospholipase A2 regulation of lipid droplet formation. Biochim. Biophys. Acta 2014, 1841, 1661–1671. [Google Scholar] [CrossRef]
- Ruipérez, V.; Astudillo, A.M.; Balboa, M.A.; Balsinde, J. Coordinate regulation of Toll-like receptor- mediated arachidonic acid mobilization in macrophages by group IVA and group V phospholipase A2s. J. Immunol. 2009, 182, 3877–3883. [Google Scholar] [CrossRef]
- Rubio, J.M.; Rodríguez, J.P.; Gil-de-Gómez, L.; Guijas, C.; Balboa, M.A.; Balsinde, J. Group V secreted phospholipase A2 is up-regulated by interleukin-4 in human macrophages and mediates phagocytosis via hydrolysis of ethanolamine phospholipids. J. Immunol. 2015, 194, 3327–3339. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, L.; Ye, X.; Chen, L.; Zhang, L.; Gao, Y.; Kang, J.X.; Cai, C. Characteristics of fatty acid distribution is associated with colorectal cancer prognosis. Prostag. Leukot. Essent. Fat. Acids 2013, 88, 355–360. [Google Scholar] [CrossRef]
- Bevers, L.E.; Pinkse, M.W.H.; Verhaert, P.D.E.M.; Hagen, W.R. Oleate hydratase catalyzes the hydration of a nonactivated carbon-carbon bond. J. Bacteriol. 2009, 191, 5010–5012. [Google Scholar] [CrossRef]
- Morvan, B.; Joblin, K.N. Hydration of oleic acid by Enterococcus gallinarum, Pediococcus acidilactici and Lactobacillus sp. isolated from the rumen. Anaerobe 1999, 5, 605–611. [Google Scholar] [CrossRef]
- Wilson, R.; Lyall, K.; Smyth, L.; Fernie, C.E.; Riemersma, R.A. Dietary hydroxy fatty acids are absorbed in humans: Implications for the measurement of “oxidative stress” in vivo. Free Radic. Biol. Med. 2002, 32, 162–168. [Google Scholar] [CrossRef]
- Kuda, O.; Brezinova, M.; Silhavy, J.; Landa, V.; Zidek, V.; Dodia, C.; Kreuchwig, F.; Vrbacky, M.; Balas, L.; Durand, T.; et al. Nrf2-mediated antioxidant defense and peroxiredoxin 6 are linked to biosynthesis of palmitic acid ester of 9-hydroxystearic acid. Diabetes 2018, 67, 1190–1199. [Google Scholar] [CrossRef]
- Balas, L.; Feillet-Coudray, C.; Durand, T. Branched fatty acyl esters of hydroxyl fatty acids, appealing beneficial endogenous fat against obesity and type-2 diabetes. Chem. Eur. J. 2018, 24, 9463–9476. [Google Scholar] [CrossRef]
- Nelson, A.T.; Kolar, M.J.; Chu, Q.; Syed, I.; Kahn, B.B.; Saghatelian, A.; Siegel, D. Stereochemistry of endogenous palmitic acid ester of 9-hydroxystearic acid and relevance of absolute configuration to regulation. J. Am. Chem. Soc. 2017, 139, 4943–4947. [Google Scholar] [CrossRef]
- Boulares, A.H.; Yakovlev, A.G.; Ivanova, V.; Stoica, B.A.; Wang, G.; Iyer, S.; Smulson, M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase-3-resistant PARP mutant increases rates of apoptosis in transfected cells. J. Biol. Chem. 1999, 274, 22932–22940. [Google Scholar] [CrossRef]
- Pérez, R.; Matabosch, X.; Llebaria, A.; Balboa, M.A.; Balsinde, J. Blockade of arachidonic acid incorporation into phospholipids induces apoptosis in U937 promonocytic cells. J. Lipid Res. 2006, 47, 484–491. [Google Scholar] [CrossRef]
- Valdearcos, M.; Esquinas, E.; Meana, C.; Peña, L.; Gil-de-Gómez, L.; Balsinde, J.; Balboa, M.A. Lipin-2 reduces proinflammatory signaling induced by saturated fatty acids in macrophages. J. Biol. Chem. 2012, 287, 10894–10904. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Diez, E.; Balsinde, J.; Aracil, M.; Schüller, A. Ethanol induces release of arachidonic acid but not synthesis of eicosanoids in mouse peritoneal macrophages. Biochim. Biophys. Acta 1987, 921, 82–89. [Google Scholar] [CrossRef]
- Astudillo, A.M.; Pérez-Chacón, G.; Balgoma, D.; Gil-de-Gómez, L.; Ruipérez, V.; Guijas, C.; Balboa, M.A.; Balsinde, J. Influence of cellular arachidonic acid levels on phospholipid remodeling and CoA-independent transacylase activity in human monocytes and U937 cells. Biochim. Biophys. Acta 2011, 1811, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Guijas, C.; Pérez-Chacón, G.; Astudillo, A.M.; Rubio, J.M.; Gil-de-Gómez, L.; Balboa, M.A.; Balsinde, J. Simultaneous activation of p38 and JNK by arachidonic acid stimulates the cytosolic phospholipase A2-dependent synthesis of lipid droplets in human monocytes. J. Lipid Res. 2012, 53, 2343–2354. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Meana, C.; Astudillo, A.M.; Balboa, M.A.; Balsinde, J. Foamy monocytes are enriched in cis-7-hexadecenoic fatty acid, 16:1n-9, a possible biomarker for early detection of cardiovascular disease. Cell Chem. Biol. 2016, 23, 689–699. [Google Scholar] [CrossRef]
- Astudillo, A.M.; Meana, C.; Guijas, C.; Pereira, L.; Lebrero, P.; Balboa, M.A.; Balsinde, J. Occurrence and biological activity of palmitoleic acid isomers in phagocytic cells. J. Lipid Res. 2018, 59, 237–249. [Google Scholar] [CrossRef]
- Balgoma, D.; Astudillo, A.M.; Pérez-Chacón, G.; Montero, O.; Balboa, M.A.; Balsinde, J. Markers of monocyte activation revealed by lipidomic profiling of arachidonic acid-containing phospholipids. J. Immunol. 2010, 184, 3857–3865. [Google Scholar] [CrossRef] [PubMed]
- Astudillo, A.M.; Pérez-Chacón, G.; Meana, C.; Balgoma, D.; Pol, A.; del Pozo, M.A.; Balboa, M.A.; Balsinde, J. Altered arachidonate distribution in macrophages from caveolin-1 null mice leading to reduced eicosanoid synthesis. J. Biol. Chem. 2011, 286, 35299–35307. [Google Scholar] [CrossRef]
- Valdearcos, M.; Esquinas, E.; Meana, C.; Gil-de-Gómez, L.; Guijas, C.; Balsinde, J.; Balboa, M.A. Subcellular localization and role of lipin-1 in human macrophages. J. Immunol. 2011, 186, 6004–6013. [Google Scholar] [CrossRef]
- Gil-de-Gómez, L.; Astudillo, A.M.; Meana, C.; Rubio, J.M.; Guijas, C.; Balboa, M.A.; Balsinde, J. A phosphatidylinositol species acutely generated by activated macrophages regulates innate immune responses. J. Immunol. 2013, 190, 5169–5177. [Google Scholar] [CrossRef]
- Gil-de-Gómez, L.; Astudillo, A.M.; Guijas, C.; Magrioti, V.; Kokotos, G.; Balboa, M.A.; Balsinde, J. Cytosolic group IVA and calcium-independent group VIA phospholipase A2s act on distinct phospholipid pools in zymosan-stimulated mouse peritoneal macrophages. J. Immunol. 2014, 192, 752–762. [Google Scholar] [CrossRef]
- Gil-de-Gómez, L.; Astudillo, A.M.; Lebrero, P.; Balboa, M.A.; Balsinde, J. Essential role for ethanolamine plasmalogen hydrolysis in bacterial lipopolysaccharide priming of macrophages for enhanced arachidonic acid release. Front. Immunol. 2017, 8, 1251. [Google Scholar] [CrossRef]
- Rubio, J.M.; Astudillo, A.M.; Casas, J.; Balboa, M.A.; Balsinde, J. Regulation of phagocytosis in macrophages by membrane ethanolamine plasmalogens. Front. Immunol. 2018, 9, 1723. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, S.; Syed, I.; Stáhlman, M.; Kolar, M.J.; Homan, E.A.; Chu, Q.; Smith, U.; Borén, J.; Kahn, B.B.; et al. A LC-MS–based workflow for measurement of branched fatty acid esters of hydroxy fatty acids. Nat. Protoc. 2016, 11, 747–763. [Google Scholar] [CrossRef]









© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, J.P.; Guijas, C.; Astudillo, A.M.; Rubio, J.M.; Balboa, M.A.; Balsinde, J. Sequestration of 9-Hydroxystearic Acid in FAHFA (Fatty Acid Esters of Hydroxy Fatty Acids) as a Protective Mechanism for Colon Carcinoma Cells to Avoid Apoptotic Cell Death. Cancers 2019, 11, 524. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11040524
Rodríguez JP, Guijas C, Astudillo AM, Rubio JM, Balboa MA, Balsinde J. Sequestration of 9-Hydroxystearic Acid in FAHFA (Fatty Acid Esters of Hydroxy Fatty Acids) as a Protective Mechanism for Colon Carcinoma Cells to Avoid Apoptotic Cell Death. Cancers. 2019; 11(4):524. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11040524
Chicago/Turabian StyleRodríguez, Juan P., Carlos Guijas, Alma M. Astudillo, Julio M. Rubio, María A. Balboa, and Jesús Balsinde. 2019. "Sequestration of 9-Hydroxystearic Acid in FAHFA (Fatty Acid Esters of Hydroxy Fatty Acids) as a Protective Mechanism for Colon Carcinoma Cells to Avoid Apoptotic Cell Death" Cancers 11, no. 4: 524. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11040524