Metabolic Plasticity in Ovarian Cancer Stem Cells
Abstract
:1. Introduction
2. Material and Methods
2.1. Microarray Extraction
2.2. Data Analysis
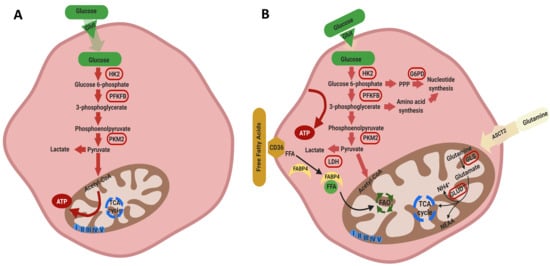
3. Results and Discussion
3.1. Correlation Between Putative OCSCs Markers
3.2. Correlation between OCSCs Markers and Glycolysis
3.3. Correlation between OCSCs Markers and TCA Cycle
3.4. OXPHOS in Ovarian Cancer Stem Cells
3.5. Correlation of OCSC Markers With Lipid Metabolism
3.6. Glutamine/Glutamate Metabolism in OCSCs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Borley, J.; Wilhelm-Benartzi, C.; Brown, R.; Ghaem-Maghami, S. Does tumour biology determine surgical success in the treatment of epithelial ovarian cancer? A systematic literature review. Br. J. Cancer 2012, 107, 1069–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Alem, L.F.; Pandya, U.M.; Baker, A.T.; Bellio, C.; Zarrella, B.D.; Clark, J.; DiGloria, C.M.; Rueda, B.R. Ovarian cancer stem cells: What progress have we made? Int. J. Biochem. Cell Biol. 2019, 107, 92–103. [Google Scholar] [CrossRef]
- Kenda Suster, N.; Virant-Klun, I. Presence and role of stem cells in ovarian cancer. World J. Stem Cells 2019, 11, 383–397. [Google Scholar] [CrossRef]
- Malanchi, I.; Santamaria-Martinez, A.; Susanto, E.; Peng, H.; Lehr, H.A.; Delaloye, J.F.; Huelsken, J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011, 481, 85–89. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells: Current status and evolving complexities. Cell Stem 2012, 10, 717–728. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Dong, J.; Haiech, J.; Kilhoffer, M.C.; Zeniou, M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016, 2016, 1740936. [Google Scholar] [CrossRef] [Green Version]
- Takeishi, S.; Nakayama, K.I. To wake up cancer stem cells, or to let them sleep, that is the question. Cancer Sci. 2016, 107, 875–881. [Google Scholar] [CrossRef]
- Zeuner, A. The secret life of quiescent cancer stem cells. Mol. Cell Oncol. 2015, 2, e968067. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.Q.; Choi, Y.P.; Kang, S.; Youn, J.H.; Cho, N.H. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene 2010, 29, 2672–2680. [Google Scholar] [CrossRef] [Green Version]
- Cheung, T.H.; Rando, T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Lin, S.Y. Tumor dormancy: Potential therapeutic target in tumor recurrence and metastasis prevention. Exp. Hematol. Oncol. 2013, 2, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sosa, M.S.; Bragado, P.; Aguirre-Ghiso, J.A. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer 2014, 14, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Brooks, M.; Wicha, M.S. Epithelial-mesenchymal plasticity of breast cancer stem cells: Implications for metastasis and therapeutic resistance. Curr. Pharm. Des. 2015, 21, 1301–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Outschoorn, U.E.; Peiris-Pages, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- Sancho, P.; Barneda, D.; Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 2016, 114, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl.) 2015, 3, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Janiszewska, M.; Suva, M.L.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clement-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef] [Green Version]
- Sancho, P.; Burgos-Ramos, E.; Tavera, A.; Bou Kheir, T.; Jagust, P.; Schoenhals, M.; Barneda, D.; Sellers, K.; Campos-Olivas, R.; Grana, O.; et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015, 22, 590–605. [Google Scholar] [CrossRef] [Green Version]
- Cantor, J.R.; Sabatini, D.M. Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2012, 2, 881–898. [Google Scholar] [CrossRef] [Green Version]
- Peixoto, J.; Lima, J. Metabolic traits of cancer stem cells. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, C.Y.; Patten, D.A.; Richardson, R.B.; Harper, M.E.; Tsang, B.K. Tumor metabolism regulating chemosensitivity in ovarian cancer. Genes Cancer 2018, 9, 155–175. [Google Scholar] [PubMed] [Green Version]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Mak, A.B.; Schnegg, C.; Lai, C.Y.; Ghosh, S.; Yang, M.H.; Moffat, J.; Hsu, M.Y. CD133-targeted niche-dependent therapy in cancer: A multipronged approach. Am. J. Pathol. 2014, 184, 1256–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florek, M.; Haase, M.; Marzesco, A.M.; Freund, D.; Ehninger, G.; Huttner, W.B.; Corbeil, D. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 2005, 319, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Hadjimichael, C.; Chanoumidou, K.; Papadopoulou, N.; Arampatzi, P.; Papamatheakis, J.; Kretsovali, A. Common stemness regulators of embryonic and cancer stem cells. World J. Stem Cells 2015, 7, 1150–1184. [Google Scholar]
- Tomita, H.; Tanaka, K.; Tanaka, T.; Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016, 7, 11018–11032. [Google Scholar] [CrossRef] [Green Version]
- Bapat, S.A.; Mali, A.M.; Koppikar, C.B.; Kurrey, N.K. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005, 65, 3025–3029. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.; Stenvers, K.L. Getting to know ovarian cancer ascites: Opportunities for targeted therapy-based translational research. Front Oncol. 2013, 3, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, N.; Abubaker, K.; Findlay, J.K. Ovarian cancer stem cells: Molecular concepts and relevance as therapeutic targets. Mol. Aspects Med. 2014, 39, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Alvero, A.B.; Fu, H.H.; Holmberg, J.; Visintin, I.; Mor, L.; Marquina, C.C.; Oidtman, J.; Silasi, D.A.; Mor, G. Stem-like ovarian cancer cells can serve as tumor vascular progenitors. Stem Cells 2009, 27, 2405–2413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elzarkaa, A.A.; Sabaa, B.E.; Abdelkhalik, D.; Mansour, H.; Melis, M.; Shaalan, W.; Farouk, M.; Malik, E.; Soliman, A.A. Clinical relevance of CD44 surface expression in advanced stage serous epithelial ovarian cancer: A prospective study. J. Cancer Res. Clin. Oncol. 2016, 142, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, C.; Rouzier, R.; Geyl, C.; Cortez, A.; Castela, M.; Lis, R.; Darai, E.; Touboul, C. Predictive markers of chemoresistance in advanced stages epithelial ovarian carcinoma. Gynecol. Oncol. 2015, 136, 112–120. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Peyrollier, K.; Xia, W.; Gilad, E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J. Biol. Chem. 2008, 283, 17635–17651. [Google Scholar] [CrossRef] [Green Version]
- Klemba, A.; Purzycka-Olewiecka, J.K.; Wcislo, G.; Czarnecka, A.M.; Lewicki, S.; Lesyng, B.; Szczylik, C.; Kieda, C. Surface markers of cancer stem-like cells of ovarian cancer and their clinical relevance. Contemp. Oncol. (Pozn) 2018, 22, 48–55. [Google Scholar] [CrossRef]
- Abelson, S.; Shamai, Y.; Berger, L.; Skorecki, K.; Tzukerman, M. Niche-dependent gene expression profile of intratumoral heterogeneous ovarian cancer stem cell populations. PLoS ONE 2013, 8, e83651. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Yang, L.; Lai, D. KLF5 strengthens drug resistance of ovarian cancer stem-like cells by regulating survivin expression. Cell Prolif. 2013, 46, 425–435. [Google Scholar] [CrossRef]
- Zhang, S.; Cui, B.; Lai, H.; Liu, G.; Ghia, E.M.; Widhopf, G.F.; Zhang, Z.; Wu, C.C.; Chen, L.; Wu, R.; et al. Ovarian cancer stem cells express ROR1, which can be targeted for anti-cancer-stem-cell therapy. Proc. Natl. Acad. Sci. USA 2014, 111, 17266–17271. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.C.; Birkbak, N.J.; Culhane, A.C.; Drapkin, R.; Fatima, A.; Tian, R.; Schwede, M.; Alsop, K.; Daniels, K.E.; Piao, H.; et al. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clin. Cancer Res. 2012, 18, 5806–5815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, T.; Sato, A.; Ohata, H.; Ikarashi, Y.; Takahashi, R.U.; Ochiya, T.; Yoshida, M.; Tsuda, H.; Onda, T.; Kato, T.; et al. Establishment and Characterization of an In Vitro Model of Ovarian Cancer Stem-like Cells with an Enhanced Proliferative Capacity. Cancer Res. 2016, 76, 150–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic. Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y.; Itano, N.; Narimatsu, H.; Kudo, T.; Hirohashi, S.; Ochiai, A.; Tohnai, I.; Ueda, M.; Kimata, K. CD44 variant exon 6 expressions in colon cancer assessed by quantitative analysis using real time reverse transcriptase-polymerase chain reaction. Oncol. Rep. 2003, 10, 1919–1924. [Google Scholar] [CrossRef] [PubMed]
- Afify, A.M.; Craig, S.; Paulino, A.F.; Stern, R. Expression of hyaluronic acid and its receptors, CD44s and CD44v6, in normal, hyperplastic, and neoplastic endometrium. Ann. Diagn. Pathol. 2005, 9, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Sosulski, A.; Horn, H.; Zhang, L.; Coletti, C.; Vathipadiekal, V.; Castro, C.M.; Birrer, M.J.; Nagano, O.; Saya, H.; Lage, K.; et al. CD44 Splice Variant v8-10 as a Marker of Serous Ovarian Cancer Prognosis. PLoS ONE 2016, 11, e0156595. [Google Scholar] [CrossRef]
- Tjhay, F.; Motohara, T.; Tayama, S.; Narantuya, D.; Fujimoto, K.; Guo, J.; Sakaguchi, I.; Honda, R.; Tashiro, H.; Katabuchi, H. CD44 variant 6 is correlated with peritoneal dissemination and poor prognosis in patients with advanced epithelial ovarian cancer. Cancer Sci. 2015, 106, 1421–1428. [Google Scholar] [CrossRef] [Green Version]
- Tamada, M.; Nagano, O.; Tateyama, S.; Ohmura, M.; Yae, T.; Ishimoto, T.; Sugihara, E.; Onishi, N.; Yamamoto, T.; Yanagawa, H.; et al. Modulation of Glucose Metabolism by CD44 Contributes to Antioxidant Status and Drug Resistance in Cancer Cells. Cancer Res. 2012, 72, 1438–1448. [Google Scholar] [CrossRef] [Green Version]
- Palorini, R.; Votta, G.; Balestrieri, C.; Monestiroli, A.; Olivieri, S.; Vento, R.; Chiaradonna, F. Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J. Cell Biochem. 2014, 115, 368–379. [Google Scholar] [CrossRef]
- Schieber, M.S.; Chandel, N.S. ROS links glucose metabolism to breast cancer stem cell and EMT phenotype. Cancer Cell 2013, 23, 265–267. [Google Scholar] [CrossRef] [Green Version]
- Penkert, J.; Ripperger, T.; Schieck, M.; Schlegelberger, B.; Steinemann, D.; Illig, T. On metabolic reprogramming and tumor biology: A comprehensive survey of metabolism in breast cancer. Oncotarget 2016, 7, 67626–67649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, D.; Ma, Y.; Liu, J.; Trope, C.G.; Holm, R.; Nesland, J.M.; Suo, Z. The hypoxic microenvironment upgrades stem-like properties of ovarian cancer cells. BMC Cancer 2012, 12, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.C.; Kim, J.H. Cancer stem cell metabolism: Target for cancer therapy. BMB Rep. 2018, 51, 319–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef] [Green Version]
- Green, D.R.; Galluzzi, L.; Kroemer, G. Cell biology. Metabolic control of cell death. Science 2014, 345, 1250256. [Google Scholar] [CrossRef] [Green Version]
- Marino, G.; Pietrocola, F.; Eisenberg, T.; Kong, Y.; Malik, S.A.; Andryushkova, A.; Schroeder, S.; Pendl, T.; Harger, A.; Niso-Santano, M.; et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell 2014, 53, 710–725. [Google Scholar] [CrossRef] [Green Version]
- Liao, J.; Qian, F.; Tchabo, N.; Mhawech-Fauceglia, P.; Beck, A.; Qian, Z.; Wang, X.; Huss, W.J.; Lele, S.B.; Morrison, C.D.; et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS ONE 2014, 9, e84941. [Google Scholar] [CrossRef] [Green Version]
- Bonuccelli, G.; De Francesco, E.M.; de Boer, R.; Tanowitz, H.B.; Lisanti, M.P. NADH autofluorescence, a new metabolic biomarker for cancer stem cells: Identification of Vitamin C and CAPE as natural products targeting “stemness”. Oncotarget 2017, 8, 20667–20678. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.P.; Liao, J.; Tang, Z.J.; Wu, W.J.; Yang, J.; Zeng, Z.L.; Hu, Y.; Wang, P.; Ju, H.Q.; Xu, R.H.; et al. Metabolic regulation of cancer cell side population by glucose through activation of the Akt pathway. Cell Death Differ. 2014, 21, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Ryan, D.G.; Murphy, M.P.; Frezza, C.; Prag, H.A.; Chouchani, E.T.; O’Neill, L.A.; Mills, E.L. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 2019, 1, 16–33. [Google Scholar] [CrossRef] [PubMed]
- Pasto, A.; Bellio, C.; Pilotto, G.; Ciminale, V.; Silic-Benussi, M.; Guzzo, G.; Rasola, A.; Frasson, C.; Nardo, G.; Zulato, E.; et al. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget 2014, 5, 4305–4319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dar, S.; Chhina, J.; Mert, I.; Chitale, D.; Buekers, T.; Kaur, H.; Giri, S.; Munkarah, A.; Rattan, R. Bioenergetic Adaptations in Chemoresistant Ovarian Cancer Cells. Sci. Rep. 2017, 7, 8760. [Google Scholar] [CrossRef]
- Sato, M.; Kawana, K.; Adachi, K.; Fujimoto, A.; Yoshida, M.; Nakamura, H.; Nishida, H.; Inoue, T.; Taguchi, A.; Takahashi, J.; et al. Spheroid cancer stem cells display reprogrammed metabolism and obtain energy by actively running the tricarboxylic acid (TCA) cycle. Oncotarget 2016, 7, 33297–33305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.Y. Therapeutic targeting of lipid synthesis metabolism for selective elimination of cancer stem cells. Arch. Pharm. Res. 2019, 42, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, J.; Chen, D.; Yang, J.; Yang, C.; Zhang, Y.; Zhang, H.; Dou, J. Evaluation of characteristics of CD44+CD117+ ovarian cancer stem cells in three dimensional basement membrane extract scaffold versus two dimensional monocultures. BMC Cell Biol. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Condello, S.; Thomes-Pepin, J.; Ma, X.; Xia, Y.; Hurley, T.D.; Matei, D.; Cheng, J.X. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 2017, 20, 303–314.e5. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Wang, J.; Zhang, L.; Wu, D.; Yu, D.; Tian, X.; Liu, J.; Jiang, X.; Shen, Y.; Zhang, L.; et al. Expressions of fatty acid synthase and HER2 are correlated with poor prognosis of ovarian cancer. Med. Oncol. 2015, 32, 391. [Google Scholar] [CrossRef] [Green Version]
- Bauerschlag, D.O.; Maass, N.; Leonhardt, P.; Verburg, F.A.; Pecks, U.; Zeppernick, F.; Morgenroth, A.; Mottaghy, F.M.; Tolba, R.; Meinhold-Heerlein, I.; et al. Fatty acid synthase overexpression: Target for therapy and reversal of chemoresistance in ovarian cancer. J. Transl. Med. 2015, 13, 146. [Google Scholar] [CrossRef] [Green Version]
- Colbert, C.L.; Kim, C.W.; Moon, Y.A.; Henry, L.; Palnitkar, M.; McKean, W.B.; Fitzgerald, K.; Deisenhofer, J.; Horton, J.D.; Kwon, H.J. Crystal structure of Spot 14, a modulator of fatty acid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 18820–18825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goren, M.A.; Fox, B.G. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr. Purif. 2008, 62, 171–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Q.; Zeng, F.; Liu, X.; Wang, Q.J.; Deng, F. Fatty acid oxidation and carnitine palmitoyltransferase I: Emerging therapeutic targets in cancer. Cell Death Dis. 2016, 7, e2226. [Google Scholar] [CrossRef] [PubMed]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 2017, 3, 169–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oburoglu, L.; Tardito, S.; Fritz, V.; de Barros, S.C.; Merida, P.; Craveiro, M.; Mamede, J.; Cretenet, G.; Mongellaz, C.; An, X.; et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell 2014, 15, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Lee, K.J.; Seo, Y.; Kwon, J.H.; Yoon, J.P.; Kang, J.Y.; Lee, H.J.; Park, S.J.; Hong, S.P.; Cheon, J.H.; et al. Effects of metformin on colorectal cancer stem cells depend on alterations in glutamine metabolism. Sci. Rep. 2018, 8, 409. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [Green Version]
- Estrela, J.M.; Ortega, A.; Obrador, E. Glutathione in cancer biology and therapy. Crit. Rev. Clin. Lab. Sci. 2006, 43, 143–181. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Moss, T.; Mangala, L.S.; Marini, J.; Zhao, H.; Wahlig, S.; Armaiz-Pena, G.; Jiang, D.; Achreja, A.; Win, J.; et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol. 2014, 10, 728. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parte, S.C.; Batra, S.K.; Kakar, S.S. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J. Ovarian Res. 2018, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.G.; Chen, C.; Yang, H.; Pan, Y.F.; Zhang, X.H. Cancer stem cell subsets and their relationships. J. Transl. Med. 2011, 9, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| TCGA | CD44 | CD24 | CD117 | CD133 | ALDH1A1 | SOX2 | 4-Oct | NANOG | NOTCH1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | |
| HK1 | 0.29 | 0.00 | 7e−04 | 0.99 | 0.15 | 0.00 | 0.14 | 0.00 | 0.13 | 0.01 | 0.03 | 0.60 | 0.03 | 0.61 | 0.06 | 0.25 | 0.40 | 0.00 |
| HK2 | 0.13 | 0.01 | 0.09 | 0.08 | 0.20 | 2.9e−05 | 0.14 | 0.00 | 0.09 | 0.07 | 0.01 | 0.82 | 0.11 | 0.03 | 0.03 | 0.60 | 0.30 | 4.9e−10 |
| SLC2A1 | 0.09 | 0.07 | 0.00 | 0.95 | 0.23 | 1.7e−06 | 0.04 | 0.42 | 0.10 | 0.03 | 0.00 | 0.87 | 0.00 | 0.95 | 0.03 | 0.60 | 0.22 | 2.9e−06 |
| SLC2A2 | 0.07 | 0.16 | −0.03 | 0.60 | 0.12 | 0.02 | −0.04 | 0.42 | 0.08 | 0.09 | −0.03 | 0.48 | −0.04 | 0.43 | 0.01 | 0.77 | 0.19 | 0.00 |
| SLC2A3 | 0.31 | 9.7e−11 | −0.02 | 0.68 | 0.19 | 0.00 | 0.00 | 0.95 | 0.07 | 0.15 | 0.11 | 0.02 | −0.03 | 0.54 | 0.03 | 0.49 | 0.24 | 3.3e−07 |
| SLC2A4 | 0.01 | 0.77 | 0.04 | 0.37 | 0.19 | 9.4e−05 | 0.01 | 0.87 | 0.00 | 0.97 | 0.09 | 0.06 | 0.01 | 0.77 | 0.09 | 0.06 | 0.18 | 0.00 |
| SLC2A5 | 0.45 | 0.00 | 0.02 | 0.74 | 0.06 | 0.24 | 0.00 | 0.98 | −0.03 | 0.61 | −0.03 | 0.57 | 0.03 | 0.52 | −0.05 | 0.29 | 0.18 | 0.00 |
| SLC2A6 | 0.24 | 5e−07 | −0.12 | 0.01 | 0.00 | 0.90 | 0.00 | 0.89 | 0.05 | 0.31 | 0.00 | 0.86 | −0.04 | 0.40 | −0.05 | 0.28 | 0.29 | 1e−09 |
| PGK1 | 0.30 | 4.4 × 1010 | −0.01 | 0.82 | 0.02 | 0.63 | 0.02 | 0.69 | 0.19 | 8.7e−05 | 0.10 | 0.04 | 0.00 | 1.00 | 0.04 | 0.45 | 0.14 | 0.00 |
| PFKFB3 | 0.35 | 0.00 | 0.00 | 0.93 | 0.11 | 0.02 | 0.11 | 0.03 | 0.04 | 0.39 | 0.10 | 0.03 | 0.09 | 0.07 | −0.01 | 0.77 | 0.28 | 6.9e−09 |
| LDHA | 0.20 | 3.5e−05 | −0.03 | 0.60 | −0.02 | 0.74 | −0.03 | 0.60 | 0.13 | 0.01 | 0.06 | 0.24 | 0.04 | 0.46 | 0.00 | 0.97 | −0.02 | 0.65 |
| FBP1 | 0.40 | 0.00 | −0.11 | 0.02 | −0.06 | 0.24 | 0.00 | 0.98 | 0.11 | 0.03 | 0.06 | 0.19 | 0.00 | 0.96 | 0.00 | 0.93 | 0.03 | 0.48 |
| PDK1 | 0.12 | 0.01 | 0.02 | 0.66 | 0.04 | 0.38 | 0.06 | 0.23 | 0.24 | 3.9e−07 | 0.04 | 0.41 | 0.04 | 0.42 | 0.01 | 0.80 | 0.13 | 0.01 |
| PDHA | 0.12 | 0.01 | −0.02 | 0.75 | −0.02 | 0.70 | 0.07 | 0.13 | 0.14 | 0.01 | 0.09 | 0.06 | 0.08 | 0.11 | 0.03 | 0.51 | 0.22 | 5.4e−06 |
| TPI1 | 0.07 | 0.15 | 0.05 | 0.33 | −0.04 | 0.47 | −0.06 | 0.25 | 0.03 | 0.56 | 0.02 | 0.75 | −0.01 | 0.77 | 0.08 | 0.10 | −0.02 | 0.74 |
| ENO1 | 0.23 | 1.3e−06 | 0.07 | 0.17 | −0.02 | 0.71 | 0.09 | 0.08 | 0.10 | 0.05 | 0.01 | 0.86 | 0.13 | 0.01 | −0.04 | 0.47 | 0.12 | 0.02 |
| PHGDH | 0.02 | 0.71 | 0.03 | 0.50 | 0.19 | 5.3e−05 | 0.10 | 0.05 | 0.20 | 4.8e−05 | 0.22 | 6.4e−06 | −0.05 | 0.30 | 0.14 | 0.00 | 0.14 | 0.00 |
| TCGA | CD44 | CD24 | CD117 | CD133 | ALDH1A1 | SOX2 | 4-Oct | NANOG | NOTCH1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | |
| CS | 0.19 | 8.2e−05 | 0.16 | 0.00 | 0.23 | 9.9e−07 | 0.09 | 0.06 | 0.15 | 0.00 | 0.00 | 0.96 | 0.07 | 0.18 | 0.11 | 0.02 | 0.39 | 0.00 |
| ACO1 | 0.30 | 1.6e−10 | 0.08 | 0.09 | 0.21 | 1.1e−05 | 0.24 | 7.6e−07 | 0.30 | 5.1e−10 | 0.10 | 0.05 | 0.11 | 0.02 | 0.00 | 0.99 | 0.34 | 2.8e−13 |
| ACO2 | 0.18 | 0.00 | 0.11 | 0.02 | −0.06 | 0.17 | 0.04 | 0.41 | −0.03 | 0.58 | −0.03 | 0.58 | 0.10 | 0.05 | −0.04 | 0.45 | 0.24 | 3.6e−07 |
| IDH1 | 0.12 | 0.01 | 0.08 | 0.11 | 0.01 | 0.77 | 0.06 | 0.21 | 0.16 | 0.00 | 0.09 | 0.08 | 0.01 | 0.88 | 0.02 | 0.63 | 0.17 | 0.00 |
| IDH2 | 0.16 | 0.00 | 0.07 | 0.13 | 0.11 | 0.03 | 0.07 | 0.14 | 0.09 | 0.08 | 0.05 | 0.27 | −0.03 | 0.51 | 0.04 | 0.39 | 0.25 | 2.6e−07 |
| IDH3A | 0.16 | 0.00 | −0.09 | 0.06 | −0.04 | 0.44 | 0.07 | 0.15 | 0.09 | 0.06 | 0.04 | 0.37 | −0.03 | 0.58 | −0.01 | 0.77 | 0.24 | 4.2e−07 |
| IDH3B | −0.08 | 0.12 | 0.09 | 0.06 | 0.09 | 0.07 | 0.04 | 0.48 | 0.03 | 0.60 | −0.05 | 0.35 | 0.06 | 0.21 | 0.04 | 0.47 | 0.24 | 6e−07 |
| SUCLG1 | 0.13 | 0.01 | 0.16 | 0.00 | 0.01 | 0.86 | 0.01 | 0.81 | 0.13 | 0.01 | 0.00 | 0.90 | 0.00 | 0.99 | 0.03 | 0.50 | 0.04 | 0.41 |
| SUCLG2 | 0.19 | 9.3e−05 | 0.01 | 0.89 | −0.08 | 0.12 | 0.05 | 0.27 | 0.06 | 0.22 | 0.08 | 0.12 | 0.11 | 0.02 | 0.00 | 0.90 | 0.05 | 0.28 |
| MDH2 | 0.20 | 4.4e−05 | 0.00 | 0.92 | −0.1 | 0.05 | 0.00 | 0.96 | 0.00 | 0.97 | 0.01 | 0.79 | 0.07 | 0.17 | 0.01 | 0.87 | 0.10 | 0.04 |
| ME2 | 0.27 | 9.6e−09 | 0.16 | 0.00 | 0.05 | 0.33 | 0.08 | 0.09 | 0.09 | 0.07 | 0.04 | 0.45 | 0.13 | 0.01 | −0.03 | 0.65 | 0.12 | 0.01 |
| TCGA | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD44 | CD24 | CD117 | CD133 | ALDH1A1 | SOX2 | 4-Oct | NANOG | NOTCH1 | ||||||||||
| Complex I | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value |
| NDUFA10 | 0.04 | 0.44 | 0.02 | 0.72 | −0.02 | 0.72 | −0.03 | 0.52 | 0.09 | 0.06 | −0.02 | 0.62 | −0.03 | 0.53 | 0.00 | 0.93 | 0.10 | 0.05 |
| NDUFA11 | −0.02 | 0.62 | 0.08 | 0.10 | 0.00 | 0.93 | −0.05 | 0.29 | −0.01 | 0.86 | −0.02 | 0.67 | −0.07 | 0.16 | 0.05 | 0.30 | −0.15 | 0.00 |
| NDUFAF4 | −0.12 | 0.01 | 0.23 | 1.6e−06 | −0.06 | 0.22 | −0.09 | 0.05 | 0.03 | 0.54 | −0.02 | 0.76 | 0.00 | 1.00 | 0.05 | 0.29 | −0.13 | 0.01 |
| NDUFA2 | −0.13 | 0.01 | −0.03 | 0.48 | −0.19 | 1e−04 | −0.11 | 0.02 | −0.02 | 0.66 | −0.02 | 0.64 | −0.09 | 0.06 | 0.01 | 0.83 | −0.29 | 1.7e−09 |
| NDUFB7 | −0.12 | 0.01 | −0.02 | 0.67 | −0.15 | 0.00 | −0.09 | 0.06 | −0.05 | 0.32 | −0.04 | 0.40 | −0.07 | 0.15 | 0.03 | 0.49 | −0.18 | 0.00 |
| NDUFS1 | 0.20 | 2.8e−05 | 0.16 | 0.00 | 0.14 | 0.00 | 0.14 | 0.00 | 0.17 | 0.00 | 0.02 | 0.69 | 0.07 | 0.14 | 0.03 | 0.57 | 0.39 | 0.00 |
| NDUFS2 | 0.18 | 0.00 | 0.13 | 0.01 | 0.15 | 0.00 | 0.13 | 0.01 | 0.06 | 0.23 | 0.09 | 0.06 | 0.15 | 0.00 | 0.16 | 0.00 | 0.36 | 1.3e−14 |
| NDUFS3 | 0.12 | 0.01 | −0.05 | 0.29 | −0.12 | 0.01 | −0.02 | 0.74 | 0.00 | 0.93 | 0.04 | 0.39 | 0.04 | 0.39 | 0.02 | 0.62 | −0.10 | 0.05 |
| NDUFS7 | 0.15 | 0.00 | −0.02 | 0.73 | −0.18 | 0.00 | −0.08 | 0.10 | −0.03 | 0.49 | −0.01 | 0.89 | −0.01 | 0.86 | −0.02 | 0.69 | −0.11 | 0.02 |
| NDUFV1 | 0.05 | 0.33 | −0.02 | 0.71 | −0.13 | 0.01 | −0.01 | 0.80 | −0.04 | 0.43 | 0.03 | 0.51 | 0.18 | 0.00 | 0.03 | 0.61 | 0.08 | 0.11 |
| NDUFV2 | −0.02 | 0.71 | 0.05 | 0.29 | −0.11 | 0.02 | 0.15 | 0.00 | 0.02 | 0.65 | 0.06 | 0.18 | −1e−04 | 1.00 | 0.05 | 0.35 | 0.01 | 0.88 |
| NDUFV3 | 0.06 | 0.24 | −0.01 | 0.84 | −0.03 | 0.56 | 0.07 | 0.14 | 0.09 | 0.07 | 0.03 | 0.52 | 0.10 | 0.03 | 0.08 | 0.10 | 0.15 | 0.00 |
| Complex II | ||||||||||||||||||
| SDHA | 0.11 | 0.03 | 0.03 | 0.52 | −0.04 | 0.46 | 0.05 | 0.35 | 0.03 | 0.52 | 0.11 | 0.03 | 0.06 | 0.20 | 0.04 | 0.39 | 0.16 | 0.00 |
| SDHB | 0.18 | 0.00 | 0.09 | 0.07 | −0.05 | 0.28 | 0.05 | 0.29 | −0.02 | 0.76 | −0.05 | 0.29 | 0.08 | 0.10 | −0.03 | 0.56 | 0.03 | 0.54 |
| SDHC | 0.16 | 0.00 | 0.11 | 0.03 | 0.10 | 0.04 | 0.07 | 0.15 | 0.21 | 1.4e−05 | 0.06 | 0.23 | −0.0029 | 0.95 | 0.11 | 0.03 | 0.11 | 0.02 |
| SDHD | 0.03 | 0.59 | 0.08 | 0.09 | −0.12 | 0.01 | 0.02 | 0.72 | 0.00 | 0.96 | 0.02 | 0.75 | 0.04 | 0.46 | −0.04 | 0.43 | −0.03 | 0.48 |
| Complex III | ||||||||||||||||||
| UQCRB | −0.12 | 0.02 | −0.06 | 0.19 | −0.20 | 2.6e−05 | −0.08 | 0.09 | −0.14 | 0.00 | −0.06 | 0.20 | −0.1 | 0.04 | −0.04 | 0.47 | −0.20 | 2.1e−05 |
| UQCRC1 | 0.03 | 0.54 | 0.08 | 0.09 | 0.00 | 0.94 | 0.06 | 0.25 | 0.08 | 0.09 | 0.04 | 0.39 | 0.06 | 0.20 | 0.13 | 0.01 | 0.17 | 0.00 |
| UQCRC2 | 0.21 | 1.3e−05 | 0.17 | 0.00 | 0.05 | 0.31 | 0.07 | 0.17 | 0.00 | 0.92 | −0.06 | 0.23 | 0.15 | 0.00 | 0.04 | 0.37 | 0.07 | 0.16 |
| UQCRH | −0.04 | 0.42 | −0.13 | 0.01 | −0.13 | 0.01 | −0.01 | 0.82 | −0.01 | 0.84 | 0.00 | 1.00 | −0.03 | 0.58 | −0.08 | 0.12 | −0.17 | 0.00 |
| UQCRQ | −0.05 | 0.34 | −0.06 | 0.20 | −0.17 | 0.00 | −0.06 | 0.19 | −0.03 | 0.60 | −0.02 | 0.70 | −0.01 | 0.77 | −0.04 | 0.41 | −0.31 | 3.6e−11 |
| UQCR10 | 0.04 | 0.43 | 0.15 | 0.00 | −0.1 | 0.04 | 0.03 | 0.50 | 0.00 | 0.97 | −0.02 | 0.70 | 0.06 | 0.23 | −0.02 | 0.66 | −0.11 | 0.02 |
| UQCR11 | −0.03 | 0.57 | −0.05 | 0.32 | −0.21 | 9.6e−06 | −0.10 | 0.04 | −0.07 | 0.18 | −0.03 | 0.50 | −0.1 | 0.04 | −0.03 | 0.59 | −0.29 | 8e−10 |
| CYCS | 0.09 | 0.05 | 0.07 | 0.18 | −0.06 | 0.20 | −0.04 | 0.43 | 0.09 | 0.06 | −0.02 | 0.73 | 0.02 | 0.64 | 0.03 | 0.60 | 0.11 | 0.03 |
| Complex IV | ||||||||||||||||||
| COX4I1 | 0.01 | 0.81 | 0.10 | 0.05 | −0.15 | 0.00 | 0.03 | 0.57 | −0.09 | 0.07 | −0.06 | 0.26 | 0.03 | 0.50 | −0.06 | 0.21 | −0.19 | 1e−04 |
| COX4I2 | −0.07 | 0.18 | 0.06 | 0.22 | 0.25 | 2.2e−07 | 0.01 | 0.83 | 0.17 | 0.00 | −0.01 | 0.82 | −0.08 | 0.10 | 0.16 | 0.00 | 0.15 | 0.00 |
| COX5A | 0.00 | 0.95 | −0.09 | 0.08 | −0.19 | 9.3e−05 | −0.04 | 0.42 | 0.00 | 0.95 | −0.04 | 0.43 | −0.04 | 0.42 | −0.05 | 0.31 | −0.13 | 0.01 |
| COX5B | −0.22 | 6.6e−06 | 0.10 | 0.04 | −0.18 | 0.00 | −0.07 | 0.13 | −0.09 | 0.08 | −0.02 | 0.64 | −0.02 | 0.68 | −0.04 | 0.36 | −0.27 | 1.7e−08 |
| COX6A1 | −0.1 | 0.04 | 0.03 | 0.53 | −0.11 | 0.03 | −0.06 | 0.23 | 0.07 | 0.13 | 0.01 | 0.87 | −0.10 | 0.04 | 0.06 | 0.19 | −0.12 | 0.02 |
| COX6A2 | −0.13 | 0.01 | 0.09 | 0.06 | 0.20 | 3.1e−05 | 0.00 | 0.92 | 0.14 | 0.00 | 0.02 | 0.69 | −0.07 | 0.14 | 0.24 | 3.6e−07 | 0.04 | 0.39 |
| COX6B1 | −0.06 | 0.21 | −0.03 | 0.61 | −0.02 | 0.73 | −0.06 | 0.23 | 0.07 | 0.15 | −0.01 | 0.87 | 0.01 | 0.85 | 0.07 | 0.15 | −0.19 | 0.00 |
| COX6B2 | −0.04 | 0.38 | −0.01 | 0.77 | 0.06 | 0.20 | −0.03 | 0.54 | 0.00 | 0.94 | −0.03 | 0.48 | 0.00 | 0.96 | 0.07 | 0.15 | 0.23 | 2.6e−06 |
| COX6C | −0.09 | 0.05 | −0.12 | 0.01 | −0.20 | 5e−05 | −0.032 | 0.51 | −0.13 | 0.01 | −0.04 | 0.39 | −0.12 | 0.01 | −0.01 | 0.91 | −0.12 | 0.01 |
| COX7A1 | −0.02 | 0.64 | −0.06 | 0.24 | 0.13 | 0.01 | −0.03 | 0.56 | 0.08 | 0.09 | −0.04 | 0.38 | −0.06 | 0.19 | 0.02 | 0.67 | 0.00 | 0.99 |
| COX7A2 | −0.08 | 0.09 | 0.09 | 0.05 | −0.04 | 0.40 | −0.09 | 0.06 | 0.02 | 0.74 | 0.00 | 0.93 | −0.08 | 0.09 | 0.01 | 0.87 | −0.19 | 5.8e−05 |
| COX7B | −0.02 | 0.70 | −0.01 | 0.78 | −0.19 | 1e−04 | −0.07 | 0.14 | 0.01 | 0.78 | 0.00 | 0.95 | −0.07 | 0.15 | 0.00 | 0.98 | −0.27 | 1.1e−08 |
| COX7C | −0.21 | 9.9e−06 | 0.03 | 0.55 | −0.08 | 0.09 | −0.09 | 0.08 | −0.10 | 0.04 | −0.09 | 0.06 | 0.00 | 0.96 | −0.01 | 0.84 | −0.30 | 2.8e−10 |
| Complex V | ||||||||||||||||||
| ATP5C1 | 0.03 | 0.51 | 0.08 | 0.11 | −0.06 | 0.24 | −0.06 | 0.22 | −0.02 | 0.62 | 0.01 | 0.87 | −0.05 | 0.36 | −0.05 | 0.28 | −0.07 | 0.13 |
| ATP5D | −0.02 | 0.63 | −0.01 | 0.91 | −0.22 | 7.6e−06 | −0.11 | 0.02 | −0.15 | 0.00 | −0.06 | 0.26 | 0.00 | 0.93 | −0.04 | 0.38 | −0.18 | 0.00 |
| ATP5E | −0.14 | 0.00 | 0.00 | 0.93 | −0.05 | 0.33 | −0.03 | 0.54 | 0.03 | 0.52 | 0.06 | 0.26 | −0.10 | 0.04 | 0.02 | 0.76 | −0.19 | 0.00 |
| ATP5G3 | 0.07 | 0.14 | −0.04 | 0.36 | −0.16 | 0.00 | −0.08 | 0.08 | −0.01 | 0.87 | −0.06 | 0.23 | −0.05 | 0.34 | −0.07 | 0.17 | −0.13 | 0.01 |
| ATP5J | −0.03 | 0.52 | −0.02 | 0.65 | −0.10 | 0.05 | −0.04 | 0.44 | 0.17 | 0.00 | −0.01 | 0.84 | 0.01 | 0.89 | 0.00 | 0.97 | −0.17 | 0.00 |
| ATP5O | −0.10 | 0.04 | −0.02 | 0.75 | −0.10 | 0.03 | −0.06 | 0.23 | −0.07 | 0.14 | −0.07 | 0.14 | 0.06 | 0.25 | 0.04 | 0.46 | −0.19 | 0.00 |
| ATP5S | 0.02 | 0.71 | 0.00 | 0.99 | 0.08 | 0.10 | 0.10 | 0.03 | 0.14 | 0.00 | −0.01 | 0.78 | 0.05 | 0.32 | 0.13 | 0.01 | 0.10 | 0.04 |
| TCGA | CD44 | CD24 | CD117 | CD133 | ALDH1A1 | SOX2 | 4-Oct | NANOG | NOTCH1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | |
| FABP4 | 0.04 | 0.43 | −0.04 | 0.44 | 0.05 | 0.28 | −0.07 | 0.13 | 0.05 | 0.33 | −0.04 | 0.43 | 0.00 | 0.87 | −0.06 | 0.21 | 0.07 | 0.13 |
| CD36 | 0.17 | 0.00 | −0.02 | 0.63 | 0.11 | 0.02 | −0.07 | 0.14 | 0.09 | 0.07 | 0.03 | 0.51 | −0.03 | 0.55 | 0.00 | 0.90 | 0.24 | 4e−07 |
| ACLY | 0.21 | 9e−06 | 0.13 | 0.01 | 0.37 | 3.1e−15 | 0.21 | 1.8e−05 | 0.15 | 0.00 | 0.04 | 0.43 | 0.01 | 0.85 | 0.12 | 0.02 | 0.51 | 0.00 |
| ACACA | 0.16 | 0.00 | 0.20 | 3.1e−05 | 0.19 | 0.00 | 0.22 | 4.5e−06 | 0.10 | 0.03 | 0.01 | 0.79 | 0.08 | 0.11 | 0.02 | 0.73 | 0.32 | 6.3e−12 |
| ACACB | 0.10 | 0.04 | 0.00 | 0.99 | 0.27 | 1.7e−08 | 0.11 | 0.02 | 0.13 | 0.01 | 0.03 | 0.58 | 0.10 | 0.04 | 0.12 | 0.02 | 0.40 | 0.00 |
| CPT1A | 0.12 | 0.01 | 0.03 | 0.56 | 0.11 | 0.02 | 0.16 | 0.00 | 0.09 | 0.07 | 0.06 | 0.20 | 0.09 | 0.07 | 0.00 | 0.89 | 0.35 | 7.1e−14 |
| CPT1B | 0.16 | 0.00 | −0.07 | 0.15 | −0.05 | 0.30 | −0.02 | 0.64 | 0.00 | 0.93 | 0.12 | 0.02 | 0.13 | 0.01 | 0.08 | 0.11 | 0.17 | 3e−04 |
| CPT2 | 0.19 | 7.7e−05 | 0.03 | 0.57 | 0.08 | 0.08 | 0.18 | 0.00 | 0.18 | 0.00 | 0.09 | 0.06 | 0.06 | 0.20 | 0.09 | 0.07 | 0.29 | 7.1e−10 |
| FASN | 0.08 | 0.08 | 0.06 | 0.25 | 0.10 | 0.03 | 0.05 | 0.31 | 0.05 | 0.28 | 0.00 | 0.85 | −0.02 | 0.66 | 0.02 | 0.67 | 0.42 | 0.00 |
| SCD1 | 0.27 | 1.3e−08 | 0.06 | 0.20 | 0.11 | 0.02 | 0.08 | 0.10 | 0.08 | 0.12 | 0.11 | 0.03 | −0.05 | 0.28 | −0.02 | 0.67 | 0.27 | 1.5e−08 |
| TCGA | CD44 | CD24 | CD117 | CD133 | ALDH1A1 | SOX2 | 4-Oct | NANOG | NOTCH1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | Pearson R | p-Value | |
| SLC1A1 | 0.32 | 1.2e−11 | 0.10 | 0.05 | 0.04 | 0.39 | 0.20 | 3.9e−05 | 0.12 | 0.01 | 0.15 | 0.00 | 0.18 | 0.00 | −0.04 | 0.40 | 0.09 | 0.06 |
| SLC1A2 | 0.16 | 0.00 | 0.00 | 0.93 | 0.25 | 1e−07 | 0.12 | 0.01 | 0.22 | 4.1e−06 | 0.05 | 0.31 | 0.04 | 0.44 | 0.07 | 0.13 | 0.14 | 0.00 |
| SLC1A3 | 0.20 | 4.6e−05 | 0.08 | 0.12 | 0.09 | 0.08 | 0.18 | 0.00 | −0.04 | 0.47 | 0.08 | 0.11 | 0.20 | 4.8e−05 | −0.01 | 0.82 | 0.15 | 0.00 |
| SLC1A4 | 0.27 | 1.5e−08 | −0.03 | 0.60 | 0.27 | 1.8e−08 | 0.13 | 0.01 | 0.25 | 2.3e−07 | 0.11 | 0.02 | 0.00 | 0.99 | 0.09 | 0.06 | 0.36 | 1.4e−14 |
| SLC1A5 | 0.24 | 3.7e−07 | −0.08 | 0.10 | 0.06 | 0.21 | 0.03 | 0.56 | 0.05 | 0.35 | 0.15 | 0.00 | 0.03 | 0.50 | −0.03 | 0.58 | 0.22 | 7.4e−06 |
| SLC1A6 | −0.02 | 0.64 | 0.02 | 0.76 | 0.03 | 0.53 | 0.02 | 0.69 | 0.00 | 0.96 | 0.01 | 0.78 | −0.04 | 0.38 | 0.06 | 0.21 | 0.04 | 0.40 |
| SLC1A7 | 0.09 | 0.08 | −0.06 | 0.21 | 0.43 | 0.00 | 0.25 | 2.4e−07 | 0.16 | 0.00 | −0.05 | 0.35 | −0.03 | 0.50 | −0.04 | 0.47 | 0.33 | 2e−12 |
| GLS | 0.15 | 0.00 | −0.01 | 0.86 | 0.11 | 0.02 | −0.01 | 0.86 | 0.18 | 0.00 | 0.03 | 0.49 | 0.05 | 0.30 | 0.13 | 0.01 | 0.36 | 2.8e−14 |
| GLUD1 | 0.10 | 0.03 | 0.02 | 0.73 | 0.11 | 0.03 | 0.06 | 0.20 | 0.11 | 0.02 | −0.02 | 0.68 | 0.14 | 0.00 | 0.05 | 0.35 | 0.25 | 2.2e−07 |
| GLUD2 | 0.19 | 7.8e−05 | 0.04 | 0.73 | 0.25 | 2.6e−07 | 0.05 | 0.31 | 0.26 | 4.8e−08 | 0.09 | 0.08 | 0.15 | 0.00 | 0.31 | 9.3e−11 | 0.27 | 1.3e−08 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghoneum, A.; Gonzalez, D.; Abdulfattah, A.Y.; Said, N. Metabolic Plasticity in Ovarian Cancer Stem Cells. Cancers 2020, 12, 1267. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12051267
Ghoneum A, Gonzalez D, Abdulfattah AY, Said N. Metabolic Plasticity in Ovarian Cancer Stem Cells. Cancers. 2020; 12(5):1267. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12051267
Chicago/Turabian StyleGhoneum, Alia, Daniela Gonzalez, Ammar Yasser Abdulfattah, and Neveen Said. 2020. "Metabolic Plasticity in Ovarian Cancer Stem Cells" Cancers 12, no. 5: 1267. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12051267