Genetic Engineering of Zebrafish in Cancer Research
Abstract
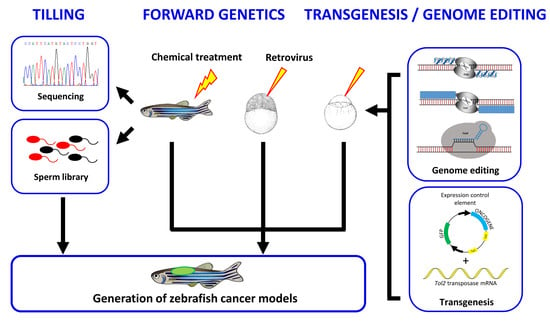
:1. Introduction
2. Chemical Carcinogenesis
3. Forward Genetics and Zebrafish Mutagenesis Screens
4. Reverse Genetics and TILLING
5. Gene Editing Technologies
5.1. Zinc Finger Nucleases
5.2. Transcription Activator-Like Effector Nucleases
5.3. Clustered Regularly Interspaced Short Palindromic Repeats RNA-Guided Cas9 Nucleases
6. Transgenesis
6.1. Tissue-Specific Promoters
6.2. The Gal4/UAS System
6.3. Cre-Mediated Recombination
6.4. Heat Shock Inducible Promoters
6.5. Tetracycline Regulated Expression
6.6. Ligand-Binding Domain Fusion Proteins
6.7. Optogenetics
6.8. Mosaic Somatic Transgenesis
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Transgenic Lines | Cancer Types | References |
|---|---|---|
| Tg(rag2: GFP-Myc) | T-ALL | [121] |
| Tg(rag2:loxP-DsRed-loxP-GFP-Myc); mRNA-Cre | [122] | |
| Tg(rag2:loxP-DsRed-loxP-GFP-Myc; hsp70:Cre) | [186] | |
| Tg(rag2:MYC-ERT2) | [208] | |
| Tg(rag2:NOTCH1ICN-GFP) | [244] | |
| Tg(rag2:GFP-Myc; rag2:GIMAP5; rag2:GIMAP7) | [245] | |
| Tg(rag2:GFP-bcl2) | T-LBL | [246] |
| Tg(rag2:GFP-Myc; rag2:GFP-bcl2) | ||
| Tg(act/xEF1:TEL-AML1)/[ETV6-RUNX1] | B-ALL | [157] |
| Tg(rag2: GFP-Myc) | [123] | |
| Tg(rag2:MYC) | [124,125] | |
| Tg(spi1:MYST3-NCOA2-GFP) [MOZ-TIF2] | AML | [131] |
| Tg(hsp70:AML1-ETO) | [181] | |
| Tg(MYCN:HSE:GFP) | [184] | |
| Tg(RUNX1-EVI1:HSE:GFP) | [185] | |
| Tg(spi1:tel-jak2a)/Tg(CMV:tel-jak2a) | [247,248] | |
| Tg(spi1:loxP-GFP-loxP-NUP98-HOXA9; hsp70:Cre) | [188] | |
| Tg(fli1:Gal4FF; UAS:GFP-HRASG12V) | [167] | |
| Tg(hsp70:p210BCR/ABL1) | [182] | |
| Tg(spi1:FLT3ITD-2A-GFP) | [132] | |
| Tg(spi1:SOX4-GFP) | [133] | |
| Tg(mitfa:BRAFV600E); tp53−/− | Melanoma | [134] |
| Tg(mitfa:BRAFV600E); mitfavc7/vc7 | [249] | |
| Tg(mitfa:GFP-NRASQ61K); tp53−/− | [135] | |
| Tg(mitfa:mitfa; mitfa:NRASQ61K); Casper | [237] | |
| Tg(mitfa:HRASG12V; mitfa:GFP) | [136] | |
| Tg(UAS:GFP-HRASG12V; mitfa/kita:Gal4-VP16) | [166,250] | |
| Tg(mitfa:GNAQQ209P); tp53−/− | [137] | |
| Tg(kita/mitfa:LexPR-Cerulean; lexOp:RFP-KrasG12V/HrasG12V/NrasQ61K) | [218] | |
| Tg(mitfa:BRAFV600E); tp53−/−; prdm1a+/− | [251] | |
| Tg(rag2:KRASG12D) | Rhabdomyosarcoma | [129] |
| Tg(cdh15:KRASG12D), Tg(mylz2:KRASG12D) | [138] | |
| Tg(ptf1a:GFP-KRASG12V) | Pancreas tumors | [139] |
| Tg(ptf1a:GAL4-VP16; UAS:KRASmut) | [170,172] | |
| Tg(myod:MYCN) | [140] | |
| Tg(ptf1a:CreERT2; ubb:loxP-STOP-loxP-Gal4; UAS:KRASG12V) | [226] | |
| Tg(ela3I:Cre; ubb:loxP-STOP-loxP-KRASG12D) | [176] | |
| Tg(act:loxP-EGFP-loxP-KRASG12D; hsp70:Cre) | Various | [187] |
| Tg(act: EWS-FLI1)/Tg(hsp70: EWS-FLI1) (tp53–/–) | Ewing sarcoma | [183] |
| Tg(rag2:myr-Akt2) | Liposarcoma | [130] |
| Tg(krt4:myr-AKT1) | [252] | |
| Tg(fabp10a:GFP-krasG12V) | Liver tumors | [141] |
| Tg(lexA:GFP-krasG12V; fapb10a:LexPR) | [215] | |
| Tg(TetO:Myc; fapb10a:rtTA) | [197,198] | |
| Tg(TetO:xmrk; fapb10a:rtTA) | [196,198] | |
| Tg(TetO:xmrk; fapb10a:rtTA; fabp10a:mCherry-T2A-twist1a-ERT2) | [210] | |
| Tg(fabp10a:HBx-RFP); tp53−/−/Tg(fabp10a:src); tp53−/− | [142,201] | |
| Tg(fabp10a:UHRF1-GFP) | [143] | |
| Tg(fabp10a:rtTA2s-M2; TRE2:EGFP-krasG12V) | [199,200,201] | |
| Tg(fabp10a:rtTA2s-M2; TRE2:EGFP-krasG12V; TRE2:EGFP-rhoAT19N) | [199] | |
| Tg(fabp10a:nrasQ61K) | [144] | |
| Tg(fabp10a:RPIA) | [145] | |
| Tg(fabp10a:LexPR; LexOp:Cre; fabp10a:loxP-mCherry-loxP-EGFP-krasG12V) | [220] | |
| Tg(fabp10a:LexPR-T2a-mcherry; LexOp:tgfb1a) | [216,217] | |
| Tg(fabp10a: AURKAV352I; myl7:EGFP) | [146] | |
| Tg(fabp10a: Xla.Ctnnb1S33A, S37A, T41A, S45A) | [147] | |
| Tg(fabp10a:CreERT2; fapb10a:loxP-BFP-loxP-Xla.Ctnnb1S33A, S37A, T41A, S45A) | [225] | |
| Tg(fapb10a:edn1) | [148] | |
| Tg(ifapb:LexPR; LexOp:EGFP-krasG12V) | Intestinal tumors | [219] |
| Tg(UAS:GFP-HRASG12V; twhh/mü4465:Gal4) | Chordoma | [169] |
| Tg(UAS:myr-AKT1; ptf1a:Gal4-VP16) | Brain tumors | [171] |
| Tg(UAS:GFP-RAC1G12V; ptf1a:Gal4-VP16) | [171] | |
| Tg(UAS:smoa1-GFP; krt5:Gal4-VP16) | [253] | |
| Tg(UAS:mCherry-KRASG12V; krt5/gfap:Gal4-VP16) | [195] | |
| Tg(TRE:mCherry-KRASG12V; krt5/gfap:rtTa) | [195] | |
| Tg(UAS:GFP-HRASG12V; zic4:Gal4-VP16) | [168] | |
| Tg(dbh:GFP-MYCN), Tg(dbh:ALKF1174L) | [149] | |
| Tg(dbh:EGFP-MYCN; dbh:LMO1) | [254] | |
| Tg(dbh:EGFP-MYCN); nf1a−/− | [155] | |
| Tg(sox10:mCherry-NRAS/NRASQ61R/NRASS17N); p53−/− | [150] | |
| Tg(dbh:MYC)/Tg(dbh:MYCN) | [151] | |
| Tg(dbh:GFP-MYCN; dbh:LIN28B) | [255] | |
| Tg(actb2:KITD816V-GFP) | Mastocytosis | [256] |
| Tg(pomc:pttg; POMC:GFP) | Pituitary adenoma | [257] |
| Tg(tg:BRAFV600E; tg:TdTomato) | Papillary thyroid carcinoma | [90] |
| Tg(flck:Tag)/Tg(flck:scl)/Tg(flck:scl; flck:lmo1) | Testicular germ cell tumors | [152] |
| Tg(ubi:loxP-EosFP-loxP-KRASG12V-T2A-mTFP; ubi:CreERT2) | Abdominal tumors | [232] |
| Tg(UAS:KRASG12V-T2A-CFP; ubi:Gal4-ERT2) |
References
- Plehn, M. Ueber geschwülste bei kaltblütern. Z. Krebs. 1906, 4, 525–564. [Google Scholar] [CrossRef] [Green Version]
- Kosswig, C. Uber bastarde der teleostier platypoecilus und xiphophorus. Z. Indukt. Abstamm. -Vererb. 1927, 44, 253. [Google Scholar]
- Kosswig, C. Uber kreuzungen zwischen den teleostiern xiphophorus helleri und platypoecilus maculatus. Z. Indukt. Abstamm. -Vererb. 1928, 47, 150–158. [Google Scholar]
- Haussler, G. Uber melanobildung bei bastarden von xiphophorus helleri und platypoecilus maculatus var. Rubra. Klin. Wochenschr. 1928, 7, 1561–1562. [Google Scholar] [CrossRef]
- Gordon, M. Hereditary basis of melanosis in hybrid fishes. Am. J. Cancer 1931, 15, 1495–1523. [Google Scholar]
- Walter, R.B.; Kazianis, S. Xiphophorus interspecies hybrids as genetic models of induced neoplasia. ILAR J. 2001, 42, 299–321. [Google Scholar] [CrossRef] [Green Version]
- Patton, E.E.; Mitchell, D.L.; Nairn, R.S. Genetic and environmental melanoma models in fish. Pigment Cell Melanoma Res. 2010, 23, 314–337. [Google Scholar] [CrossRef]
- Harshbarger, J.C.; Slatick, M.S. Lesser known aquarium fish tumor models. Mar. Biotechnol. 2001, 3, S115–S129. [Google Scholar] [CrossRef] [PubMed]
- Engeszer, R.E.; Patterson, L.B.; Rao, A.A.; Parichy, D.M. Zebrafish in the wild: A review of natural history and new notes from the field. Zebrafish 2007, 4, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Sundin, J.; Morgan, R.; Finnøen, M.H.; Dey, A.; Sarkar, K.; Jutfelt, F. On the observation of wild zebrafish (Danio rerio) in India. Zebrafish 2019, 16, 546–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laale, H.W. The biology and use of zebrafish, Brachydanio rerio in fisheries research. A literature review. J. Fish. Biol. 1977, 10, 121–173. [Google Scholar] [CrossRef]
- Grunwald, D.J.; Streisinger, G. Induction of recessive lethal and specific locus mutations in the zebrafish with ethyl nitrosourea. Genet. Res. 1992, 59, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rennekamp, A.J.; Peterson, R.T. 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 2015, 24, 58–70. [Google Scholar] [CrossRef] [Green Version]
- Dang, M.; Fogley, R.; Zon, L.I. Identifying novel cancer therapies using chemical genetics and zebrafish. Adv. Exp. Med. Biol. 2016, 916, 103–124. [Google Scholar] [CrossRef]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2008, 2, 183–189. [Google Scholar] [CrossRef] [Green Version]
- Antinucci, P.; Hindges, R. A crystal-clear zebrafish for in vivo imaging. Sci. Rep. 2016, 6, 29490. [Google Scholar] [CrossRef] [Green Version]
- Lawson, N.D.; Weinstein, B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002, 248, 307–318. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Dong, L.; Ahn, J.; Dao, D.; Hammerschmidt, M.; Chen, J.N. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev. Biol. 2007, 304, 735–744. [Google Scholar] [CrossRef] [Green Version]
- Traver, D.; Paw, B.H.; Poss, K.D.; Penberthy, W.T.; Lin, S.; Zon, L.I. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003, 4, 1238–1246. [Google Scholar] [CrossRef]
- Amatruda, J.F.; Shepard, J.L.; Stern, H.M.; Zon, L.I. Zebrafish as a cancer model system. Cancer Cell 2002, 1, 229–231. [Google Scholar] [CrossRef] [Green Version]
- Kent, M.L.; Spitsbergen, J.M.; Matthews, J.M.; Fournie, J.W.; Westerfield, M. Diseases of Zebrafish in Research Facilities. Zebrafish International Resource Center. 2016. Available online: https://zebrafish.org/wiki/health/disease_manual/start (accessed on 10 April 2020).
- Matthews, J.L. Common diseases of laboratory zebrafish. Methods Cell Biol. 2004, 77, 617–643. [Google Scholar] [CrossRef]
- Smolowitz, R.; Hanley, J.; Richmond, H. A three-year retrospective study of abdominal tumors in zebrafish maintained in an aquatic laboratory animal facility. Biol. Bull. 2002, 203, 265–266. [Google Scholar] [CrossRef] [PubMed]
- Stanton, M. Diethylnitrosamine-induced hepatic degeneration and neoplasia in the aquarium fish, Brachydanio rerio. J. Natl. Cancer Inst. 1965, 34, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Mizgireuv, I.V.; Majorova, I.G.; Gorodinskaya, V.M.; Khudoley, V.V.; Revskoy, S.Y. Carcinogenic effect of N-nitrosodimethylamine on diploid and triploid zebrafish (Danio rerio). Toxicol. Pathol. 2004, 32, 514–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizgireuv, I.V.; Revskoy, S.Y. Transplantable tumor lines generated in clonal zebrafish. Cancer Res. 2006, 66, 3120–3125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spitsbergen, J.M.; Tsai, H.-W.; Reddy, A.; Miller, T.; Arbogast, D.; Hendricks, J.D.; Bailey, G.S. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol. Pathol. 2000, 28, 716–725. [Google Scholar] [CrossRef]
- Lam, S.H.; Wu, Y.L.; Vega, V.B.; Miller, L.D.; Spitsbergen, J.; Tong, Y.; Zhan, H.; Govindarajan, K.R.; Lee, S.; Mathavan, S.; et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat. Biotechnol. 2006, 24, 73–75. [Google Scholar] [CrossRef]
- Spitsbergen, J.M.; Tsai, H.W.; Reddy, A.; Miller, T.; Arbogast, D.; Hendricks, J.D.; Bailey, G.S. Neoplasia in zebrafish (Danio rerio) treated with N-methyl-N’-nitro-N-nitrosoguanidine by three exposure routes at different developmental stages. Toxicol. Pathol. 2000, 28, 705–715. [Google Scholar] [CrossRef]
- Beckwith, L.G.; Moore, J.L.; Tsao-Wu, G.S.; Harshbarger, J.C.; Cheng, K.C. Ethylnitrosourea induces neoplasia in zebrafish (Danio rerio). Lab. Invest. 2000, 80, 379–385. [Google Scholar] [CrossRef] [Green Version]
- Lam, S.H.; Gong, Z. Modeling liver cancer using zebrafish: A comparative oncogenomics approach. Cell Cycle 2006, 5, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Zimmering, S.; Thompson, E. Comparison of rates of sex-linked recessive lethals induced by ethylnitrosourea (ENU) in postmeiotic cells of the male and oogonia of the female Drosophila. Environ. Mutagen. 1984, 6, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, I.; Ayaki, T.; Ohshima, K. Comparative studies of dose-response curves for recessive lethal mutations induced by ethylnitrosourea in spermatogonia and in spermatozoa of Drosophila melanogaster. Environ. Mutagen. 1984, 6, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.L.; Kelly, E.M.; Hunsicker, P.R.; Bangham, J.W.; Maddux, S.C.; Phipps, E.L. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. USA 1979, 76, 5818–5819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullins, M.C.; Hammerschmidt, M.; Haffter, P.; Nüsslein-Volhard, C. Large-scale mutagenesis in the zebrafish: In search of genes controlling development in a vertebrate. Curr. Biol. 1994, 4, 189–202. [Google Scholar] [CrossRef]
- Driever, W.; Solnica-Krezel, L.; Schier, A.F.; Neuhauss, S.C.; Malicki, J.; Stemple, D.L.; Stainier, D.Y.; Zwartkruis, F.; Abdelilah, S.; Rangini, Z.; et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996, 123, 37–46. [Google Scholar]
- Haffter, P.; Granato, M.; Brand, M.; Mullins, M.C.; Hammerschmidt, M.; Kane, D.A.; Odenthal, J.; van Eeden, F.J.; Jiang, Y.J.; Heisenberg, C.P.; et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 1996, 123, 1–36. [Google Scholar]
- Van Rooijen, E.; Giles, R.H.; Voest, E.E.; van Rooijen, C.; Schulte-Merker, S.; van Eeden, F.J. LRRC50, a conserved ciliary protein implicated in polycystic kidney disease. J. Am. Soc. Nephrol. 2008, 19, 1128–1138. [Google Scholar] [CrossRef] [Green Version]
- Basten, S.G.; Davis, E.E.; Gillis, A.J.; van Rooijen, E.; Stoop, H.; Babala, N.; Logister, I.; Heath, Z.G.; Jonges, T.N.; Katsanis, N.; et al. Mutations in LRRC50 predispose zebrafish and humans to seminomas. PLoS Genet. 2013, 9, e1003384. [Google Scholar] [CrossRef]
- Neumann, J.C.; Chandler, G.L.; Damoulis, V.A.; Fustino, N.J.; Lillard, K.; Looijenga, L.; Margraf, L.; Rakheja, D.; Amatruda, J.F. Mutation in the type IB bone morphogenetic protein receptor Alk6b impairs germ-cell differentiation and causes germ-cell tumors in zebrafish. Proc. Natl. Acad. Sci. USA 2011, 108, 13153–13158. [Google Scholar] [CrossRef] [Green Version]
- Amsterdam, A.; Burgess, S.; Golling, G.; Chen, W.; Sun, Z.; Townsend, K.; Farrington, S.; Haldi, M.; Hopkins, N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999, 13, 2713–2724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amsterdam, A.; Sadler, K.C.; Lai, K.; Farrington, S.; Bronson, R.T.; Lees, J.A.; Hopkins, N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004, 2, E139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amsterdam, A.; Lai, K.; Komisarczuk, A.Z.; Becker, T.S.; Bronson, R.T.; Hopkins, N.; Lees, J.A. Zebrafish hagoromo mutants up-regulate fgf8 postembryonically and develop neuroblastoma. Mol. Cancer Res. 2009, 7, 841–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shepard, J.L.; Amatruda, J.F.; Stern, H.M.; Subramanian, A.; Finkelstein, D.; Ziai, J.; Finley, K.R.; Pfaff, K.L.; Hersey, C.; Zhou, Y.; et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc. Natl. Acad. Sci. USA 2005, 102, 13194–13199. [Google Scholar] [CrossRef] [Green Version]
- Shepard, J.L.; Amatruda, J.F.; Finkelstein, D.; Ziai, J.; Finley, K.R.; Stern, H.M.; Chiang, K.; Hersey, C.; Barut, B.; Freeman, J.L.; et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007, 21, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Koudijs, M.J.; den Broeder, M.J.; Keijser, A.; Wienholds, E.; Houwing, S.; van Rooijen, E.M.; Geisler, R.; van Eeden, F.J. The zebrafish mutants dre, uki, and lep encode negative regulators of the hedgehog signaling pathway. PLoS Genet. 2005, 1, e19. [Google Scholar] [CrossRef] [Green Version]
- Niyaz, M.; Khan, M.S.; Mudassar, S. Hedgehog signaling: An achilles’ heel in cancer. Transl. Oncol. 2019, 12, 1334–1344. [Google Scholar] [CrossRef]
- Jeng, K.S.; Chang, C.F.; Lin, S.S. Sonic hedgehog signaling in organogenesis, tumors, and tumor microenvironments. Int. J. Mol. Sci. 2020, 21, 758. [Google Scholar] [CrossRef] [Green Version]
- Parant, J.M.; George, S.A.; Holden, J.A.; Yost, H.J. Genetic modeling of Li-Fraumeni syndrome in zebrafish. Dis. Model. Mech. 2010, 3, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Sonawane, M.; Carpio, Y.; Geisler, R.; Schwarz, H.; Maischein, H.M.; Nuesslein-Volhard, C. Zebrafish penner/lethal giant larvae 2 functions in hemidesmosome formation, maintenance of cellular morphology and growth regulation in the developing basal epidermis. Development 2005, 132, 3255–3265. [Google Scholar] [CrossRef] [Green Version]
- Reischauer, S.; Levesque, M.P.; Nüsslein-Volhard, C.; Sonawane, M. Lgl2 executes its function as a tumor suppressor by regulating ErbB signaling in the zebrafish epidermis. PLoS Genet. 2009, 5, e1000720. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.N.; Dolan, A.C.; Seiler, C.; Smith, E.M.; Yusuff, S.; Chaille-Arnold, L.; Judson, B.; Sierk, R.; Yengo, C.; Sweeney, H.L.; et al. Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev. Cell 2005, 8, 717–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, J.L.; Rush, L.M.; Breneman, C.; Mohideen, M.A.; Cheng, K.C. Zebrafish genomic instability mutants and cancer susceptibility. Genetics 2006, 174, 585–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazer, J.K.; Meeker, N.D.; Rudner, L.; Bradley, D.F.; Smith, A.C.; Demarest, B.; Joshi, D.; Locke, E.E.; Hutchinson, S.A.; Tripp, S.; et al. Heritable T-cell malignancy models established in a zebrafish phenotypic screen. Leukemia 2009, 23, 1825–1835. [Google Scholar] [CrossRef] [Green Version]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 2000, 18, 455–457. [Google Scholar] [CrossRef]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant. Physiol. 2000, 123, 439–442. [Google Scholar] [CrossRef] [Green Version]
- Coghill, E.L.; Hugill, A.; Parkinson, N.; Davison, C.; Glenister, P.; Clements, S.; Hunter, J.; Cox, R.D.; Brown, S.D. A gene-driven approach to the identification of ENU mutants in the mouse. Nat. Genet. 2002, 30, 255–256. [Google Scholar] [CrossRef]
- Wienholds, E.; Schulte-Merker, S.; Walderich, B.; Plasterk, R.H. Target-selected inactivation of the zebrafish rag1 gene. Science 2002, 297, 99–102. [Google Scholar] [CrossRef]
- Berghmans, S.; Murphey, R.D.; Wienholds, E.; Neuberg, D.; Kutok, J.L.; Fletcher, C.D.; Morris, J.P.; Liu, T.X.; Schulte-Merker, S.; Kanki, J.P.; et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 407–412. [Google Scholar] [CrossRef] [Green Version]
- Faucherre, A.; Taylor, G.S.; Overvoorde, J.; Dixon, J.E.; den Hertog, J. Zebrafish pten genes have overlapping and non-redundant functions in tumorigenesis and embryonic development. Oncogene 2008, 27, 1079–1086. [Google Scholar] [CrossRef] [Green Version]
- Choorapoikayil, S.; Kuiper, R.V.; de Bruin, A.; den Hertog, J. Haploinsufficiency of the genes encoding the tumor suppressor Pten predisposes zebrafish to hemangiosarcoma. Dis. Model. Mech. 2012, 5, 241–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haramis, A.P.; Hurlstone, A.; van der Velden, Y.; Begthel, H.; van den Born, M.; Offerhaus, G.J.; Clevers, H.C. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 2006, 7, 444–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feitsma, H.; Kuiper, R.V.; Korving, J.; Nijman, I.J.; Cuppen, E. Zebrafish with mutations in mismatch repair genes develop neurofibromas and other tumors. Cancer Res. 2008, 68, 5059–5066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shive, H.R.; West, R.R.; Embree, L.J.; Azuma, M.; Sood, R.; Liu, P.; Hickstein, D.D. brca2 in zebrafish ovarian development, spermatogenesis, and tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 19350–19355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurlstone, A.F.; Haramis, A.P.; Wienholds, E.; Begthel, H.; Korving, J.; Van Eeden, F.; Cuppen, E.; Zivkovic, D.; Plasterk, R.H.; Clevers, H. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature 2003, 425, 633–637. [Google Scholar] [CrossRef]
- Wimmer, K.; Rosenbaum, T.; Messiaen, L. Connections between constitutional mismatch repair deficiency syndrome and neurofibromatosis type 1. Clin. Genet. 2017, 91, 507–519. [Google Scholar] [CrossRef]
- Shive, H.R.; West, R.R.; Embree, L.J.; Golden, C.D.; Hickstein, D.D. BRCA2 and TP53 collaborate in tumorigenesis in zebrafish. PLoS ONE 2014, 9, e87177. [Google Scholar] [CrossRef] [Green Version]
- Parant, J.M.; Yeh, J.R. Approaches to inactivate genes in zebrafish. Adv. Exp. Med. Biol. 2016, 916, 61–86. [Google Scholar] [CrossRef]
- Gut, P.; Reischauer, S.; Stainier, D.Y.R.; Arnaout, R. Little fish, big data: Zebrafish as model for cardiovascular and metabolic disease. Physiol. Rev. 2017, 97, 889–938. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Noyes, B.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekker, S.C. Zinc finger-based knockout punches for zebrafish genes. Zebrafish 2008, 5, 121–123. [Google Scholar] [CrossRef]
- Shin, J.; Padmanabhan, A.; de Groh, E.D.; Lee, J.S.; Haidar, S.; Dahlberg, S.; Guo, F.; He, S.; Wolman, M.A.; Granato, M.; et al. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Dis. Model. Mech. 2012, 5, 881–894. [Google Scholar] [CrossRef] [Green Version]
- Gjini, E.; Mansour, M.R.; Sander, J.D.; Moritz, N.; Nguyen, A.T.; Kesarsing, M.; Gans, E.; He, S.; Chen, S.; Ko, M.; et al. A zebrafish model of myelodysplastic syndrome produced through tet2 genomic editing. Mol. Cell. Biol. 2015, 35, 789–804. [Google Scholar] [CrossRef] [Green Version]
- Solin, S.L.; Shive, H.R.; Woolard, K.D.; Essner, J.J.; McGrail, M. Rapid tumor induction in zebrafish by TALEN-mediated somatic inactivation of the retinoblastoma1 tumor suppressor rb1. Sci. Rep. 2015, 5, 13745. [Google Scholar] [CrossRef] [Green Version]
- Shim, J.; Choi, J.H.; Park, M.H.; Kim, H.; Kim, J.H.; Kim, S.Y.; Hong, D.; Kim, S.; Lee, J.E.; Kim, C.H.; et al. Development of zebrafish medulloblastoma-like PNET model by TALEN-mediated somatic gene inactivation. Oncotarget 2017, 8, 55280–55297. [Google Scholar] [CrossRef] [Green Version]
- Schultz, L.E.; Haltom, J.A.; Almeida, M.P.; Wierson, W.A.; Solin, S.L.; Weiss, T.J.; Helmer, J.A.; Sandquist, E.J.; Shive, H.R.; McGrail, M. Epigenetic regulators Rbbp4 and Hdac1 are overexpressed in a zebrafish model of RB1 embryonal brain tumor, and are required for neural progenitor survival and proliferation. Dis. Model. Mech. 2018, 11, dmm034124. [Google Scholar] [CrossRef] [Green Version]
- Ignatius, M.S.; Hayes, M.N.; Moore, F.E.; Tang, Q.; Garcia, S.P.; Blackburn, P.R.; Baxi, K.; Wang, L.; Jin, A.; Ramakrishnan, A.; et al. tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. Elife 2018, 7, e37202. [Google Scholar] [CrossRef]
- Jung, I.H.; Jung, D.E.; Chung, Y.Y.; Kim, K.S.; Park, S.W. Iroquois homeobox 1 acts as a true tumor suppressor in multiple organs by regulating cell cycle progression. Neoplasia 2019, 21, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Ablain, J.; Xu, M.; Rothschild, H.; Jordan, R.C.; Mito, J.K.; Daniels, B.H.; Bell, C.F.; Joseph, N.M.; Wu, H.; Bastian, B.C.; et al. Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science 2018, 362, 1055–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, M.A.; Liao, Z.W.; Yamagata, N.; Pouliot, G.P.; Stevenson, K.E.; Neuberg, D.S.; Thorner, A.R.; Ducar, M.; Silverman, E.A.; Hunger, S.P.; et al. Hedgehog pathway mutations drive oncogenic transformation in high-risk T-cell acute lymphoblastic leukemia. Leukemia 2018, 32, 2126–2137. [Google Scholar] [CrossRef] [PubMed]
- Oppel, F.; Tao, T.; Shi, H.; Ross, K.N.; Zimmerman, M.W.; He, S.; Tong, G.; Aster, J.C.; Look, A.T. Loss of atrx cooperates with p53-deficiency to promote the development of sarcomas and other malignancies. PLoS Genet. 2019, 15, e1008039. [Google Scholar] [CrossRef] [Green Version]
- Oppel, F.; Ki, D.H.; Zimmermann, M.W.; Ross, K.N.; Tao, T.; Shi, H.; He, S.; Aster, J.C.; Look, A.T. suz12 inactivation in p53 and nf1 deficient zebrafish accelerates the onset of MPNSTs and expands the spectrum of tumor types to include adenocarcinoma, leukemia, and soft tissue sarcoma. Dis. Model. Mech. 2020. Epub ahead of print. PMID: 32651197. [Google Scholar] [CrossRef]
- Shi, H.; Tao, T.; Abraham, B.J.; Durbin, A.D.; Zimmerman, M.W.; Kadoch, C.; Look, A.T. ARID1A loss in neuroblastoma promotes the adrenergic-to-mesenchymal transition by regulating enhancer-mediated gene expression. Sci. Adv. 2020, 6, eaaz3440. [Google Scholar] [CrossRef]
- Brandt, Z.J.; North, P.N.; Link, B.A. Somatic mutations of lats2 cause peripheral nerve sheath tumors in zebrafish. Cells 2019, 8, E972. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Kim, E.J.; Kim, D.H.; Park, S.W. Dact2 is involved in the regulation of epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2020, 524, 190–197. [Google Scholar] [CrossRef]
- Anelli, V.; Villefranc, J.A.; Chhangawala, S.; Martinez-McFaline, R.; Riva, E.; Nguyen, A.; Verma, A.; Bareja, R.; Chen, Z.; Scognamiglio, T.; et al. Oncogenic BRAF disrupts thyroid morphogenesis and function via twist expression. Elife 2017, 6, e20728. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011, 29, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Cade, L.; Khayter, C.; Reyon, D.; Peterson, R.T.; Joung, J.K.; Yeh, J.R. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011, 29, 697–698. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Petree, C.; Requena, T.; Varshney, P.; Varshney, G.K. Expanding the CRISPR toolbox in zebrafish for studying development and disease. Front. Cell Dev. Biol. 2019, 7, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013, 23, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [Green Version]
- Ablain, J.; Durand, E.M.; Yang, S.; Zhou, Y.; Zon, L.I. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 2015, 32, 756–764. [Google Scholar] [CrossRef] [Green Version]
- Ablain, J.; Zon, L.I. Tissue-specific gene targeting using CRISPR/Cas9. Methods Cell Biol. 2016, 135, 189–202. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, G.K.; Pei, W.; LaFave, M.C.; Idol, J.; Xu, L.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M.; et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hruscha, A.; Krawitz, P.; Rechenberg, A.; Heinrich, V.; Hecht, J.; Haass, C.; Schmid, B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 2013, 140, 4982–4987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamal, M.; Ullah, A.; Ahsan, M.; Tyagi, R.; Habib, Z.; Rehman, K. Improving CRISPR-Cas9 on-target specificity. Curr. Issues Mol. Biol. 2018, 26, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Cavodeassi, F.; Modolell, J.; Gómez-Skarmeta, J.L. The Iroquois family of genes: From body building to neural patterning. Development 2001, 128, 2847–2855. [Google Scholar]
- Liu, X.; Zhang, J.; Liu, L.; Jiang, Y.; Ji, J.; Yan, R.; Zhu, Z.; Yu, Y. Protein arginine methyltransferase 5-mediated epigenetic silencing of IRX1 contributes to tumorigenicity and metastasis of gastric cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2835–2844. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, N.; Xu, X.; Wang, Z.; Cheng, Y.; Jin, W.; Wang, X.; Yang, H.; Liu, H.; Zhang, Y.; et al. Clinical significance of Iroquois Homeobox Gene—IRX1 in human glioma. Mol. Med. Rep. 2018, 17, 4651–4656. [Google Scholar] [CrossRef] [Green Version]
- Bennett, K.L.; Karpenko, M.; Lin, M.T.; Claus, R.; Arab, K.; Dyckhoff, G.; Plinkert, P.; Herpel, E.; Smiraglia, D.; Plass, C. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008, 68, 4494–4499. [Google Scholar] [CrossRef] [Green Version]
- Stuart, G.W.; McMurray, J.V.; Westerfield, M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development 1988, 103, 403–412. [Google Scholar]
- Culp, P.; Nüsslein-Volhard, C.; Hopkins, N. High-frequency germ-line transmission of plasmid DNA sequences injected into fertilized zebrafish eggs. Proc. Natl. Acad. Sci. USA 1991, 88, 7953–7957. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, P.D.; Peek, A.; Thorgaard, G. An in vivo screen for the luciferase transgene in zebrafish. Mol. Mar. Biol. Biotechnol. 1994, 3, 307–316. [Google Scholar] [PubMed]
- Udvadia, A.J.; Linney, E. Windows into development: Historic, current, and future perspectives on transgenic zebrafish. Dev. Biol. 2003, 256, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Thermes, V.; Grabher, C.; Ristoratore, F.; Bourrat, F.; Choulika, A.; Wittbrodt, J.; Joly, J.S. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 2002, 118, 91–98. [Google Scholar] [CrossRef]
- Soroldoni, D.; Hogan, B.M.; Oates, A.C. Simple and efficient transgenesis with meganuclease constructs in zebrafish. Methods Mol. Biol. 2009, 546, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K.; Koga, A.; Hori, H.; Shima, A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene 1998, 225, 17–22. [Google Scholar] [CrossRef]
- Kawakami, K.; Shima, A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene 1999, 240, 239–244. [Google Scholar] [CrossRef]
- Kawakami, K.; Shima, A.; Kawakami, N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 2000, 97, 11403–11408. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004, 77, 201–222. [Google Scholar] [CrossRef]
- Clark, K.J.; Urban, M.D.; Skuster, K.J.; Ekker, S.C. Transgenic zebrafish using transposable elements. Methods Cell Biol. 2011, 104, 137–149. [Google Scholar] [CrossRef] [Green Version]
- Langenau, D.M.; Traver, D.; Ferrando, A.A.; Kutok, J.L.; Aster, J.C.; Kanki, J.P.; Lin, S.; Prochownik, E.; Trede, N.S.; Zon, L.I.; et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 2003, 299, 887–890. [Google Scholar] [CrossRef] [Green Version]
- Langenau, D.M.; Feng, H.; Berghmans, S.; Kanki, J.P.; Kutok, J.L.; Look, A.T. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2005, 102, 6068–6073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, E.G.; Iyer, S.; Garcia, S.P.; Loontiens, S.; Sadreyev, R.I.; Speleman, F.; Langenau, D.M. Cell of origin dictates aggression and stem cell number in acute lymphoblastic leukemia. Leukemia 2018, 32, 1860–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borga, C.; Park, G.; Foster, C.; Burroughs-Garcia, J.; Marchesin, M.; Shah, R.; Hasan, A.; Ahmed, S.T.; Bresolin, S.; Batchelor, L.; et al. Simultaneous B and T cell acute lymphoblastic leukemias in zebrafish driven by transgenic MYC: Implications for oncogenesis and lymphopoiesis. Leukemia 2019, 33, 333–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, G.; Burroughs-Garcia, J.; Foster, C.A.; Hasan, A.; Borga, C.; Frazer, J.K. Zebrafish B cell acute lymphoblastic leukemia: New findings in an old model. Oncotarget 2020, 11, 1292–1305. [Google Scholar] [CrossRef] [Green Version]
- Sinha, A.A.; Park, G.; Frazer, J.K. Tackling acute lymphoblastic leukemia-one fish at a time. Int. J. Mol. Sci. 2019, 20, 5313. [Google Scholar] [CrossRef] [Green Version]
- Jessen, J.R.; Jessen, T.N.; Vogel, S.S.; Lin, S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis 2001, 29, 156–162. [Google Scholar] [CrossRef]
- Langenau, D.M.; Ferrando, A.A.; Traver, D.; Kutok, J.L.; Hezel, J.P.; Kanki, J.P.; Zon, L.I.; Look, A.T.; Trede, N.S. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. USA 2004, 101, 7369–7374. [Google Scholar] [CrossRef] [Green Version]
- Langenau, D.M.; Keefe, M.D.; Storer, N.Y.; Guyon, J.R.; Kutok, J.L.; Le, X.; Goessling, W.; Neuberg, D.S.; Kunkel, L.M.; Zon, L.I. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007, 21, 1382–1395. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, A.; Snyder, E.L.; Marino-Enriquez, A.; Zhang, Y.X.; Sioletic, S.; Kozakewich, E.; Grebliunaite, R.; Ou, W.B.; Sicinska, E.; Raut, C.P.; et al. Aberrant AKT activation drives well-differentiated liposarcoma. Proc. Natl. Acad. Sci. USA 2011, 108, 16386–16391. [Google Scholar] [CrossRef] [Green Version]
- Zhuravleva, J.; Paggetti, J.; Martin, L.; Hammann, A.; Solary, E.; Bastie, J.N.; Delva, L. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br. J. Haematol. 2008, 143, 378–382. [Google Scholar] [CrossRef]
- Lu, J.W.; Hou, H.A.; Hsieh, M.S.; Tien, H.F.; Lin, L.I. Overexpression of FLT3-ITD driven by spi-1 results in expanded myelopoiesis with leukemic phenotype in zebrafish. Leukemia 2016, 30, 2098–2101. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Hsieh, M.S.; Hou, H.A.; Chen, C.Y.; Tien, H.F.; Lin, L.I. Overexpression of SOX4 correlates with poor prognosis of acute myeloid leukemia and is leukemogenic in zebrafish. Blood Cancer J. 2017, 7, e593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patton, E.E.; Widlund, H.R.; Kutok, J.L.; Kopani, K.R.; Amatruda, J.F.; Murphey, R.D.; Berghmans, S.; Mayhall, E.A.; Traver, D.; Fletcher, C.D.; et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 2005, 15, 249–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dovey, M.; White, R.M.; Zon, L.I. Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish 2009, 6, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, C.; Jones, M.; Walker, P.; Kamarashev, J.; Kelly, A.; Hurlstone, A.F. Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis. Model. Mech. 2009, 2, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Mouti, M.A.; Dee, C.; Coupland, S.E.; Hurlstone, A.F. Minimal contribution of ERK1/2-MAPK signalling towards the maintenance of oncogenic GNAQQ209P-driven uveal melanomas in zebrafish. Oncotarget 2016, 7, 39654–39670. [Google Scholar] [CrossRef] [Green Version]
- Storer, N.Y.; White, R.M.; Uong, A.; Price, E.; Nielsen, G.P.; Langenau, D.M.; Zon, L.I. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogeneexpression during myogenesis. Development 2013, 140, 3040–3050. [Google Scholar] [CrossRef] [Green Version]
- Park, S.W.; Davison, J.M.; Rhee, J.; Hruban, R.H.; Maitra, A.; Leach, S.D. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology 2008, 134, 2080–2090. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.W.; Kutok, J.L.; Lee, N.H.; Piao, H.Y.; Fletcher, C.D.; Kanki, J.P.; Look, A.T. Targeted expression of human MYCN selectively causes pancreatic neuroendocrine tumors in transgenic zebrafish. Cancer Res. 2004, 64, 7256–7262. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.; Spitsbergen, J.M.; Lam, S.H.; Mathavan, S.; Parinov, S.; Gong, Z. A high level of liver-specific expression of oncogenic Kras(V12) drives robust liver tumorigenesis in transgenic zebrafish. Dis. Model. Mech. 2011, 4, 801–813. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.W.; Yang, W.Y.; Tsai, S.M.; Lin, Y.M.; Chang, P.H.; Chen, J.R.; Wang, H.D.; Wu, J.L.; Jin, S.L.; Yuh, C.H. Liver-specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PLoS ONE 2013, 8, e76951. [Google Scholar] [CrossRef] [PubMed]
- Mudbhary, R.; Hoshida, Y.; Chernyavskaya, Y.; Jacob, V.; Villanueva, A.; Fiel, M.I.; Chen, X.; Kojima, K.; Thung, S.; Bronson, R.T.; et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 2014, 25, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Leng, X.; Wang, G.; Wan, X.; Cao, H. The construction of intrahepatic cholangiocarcinoma model in zebrafish. Sci. Rep. 2017, 7, 13419. [Google Scholar] [CrossRef] [Green Version]
- Chou, Y.T.; Chen, L.Y.; Tsai, S.L.; Tu, H.C.; Lu, J.W.; Ciou, S.C.; Wang, H.D.; Yuh, C.H. Ribose-5-phosphate isomerase A overexpression promotes liver cancer development in transgenic zebrafish via activation of ERK and β-catenin pathways. Carcinogenesis 2019, 40, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.L.; Su, C.W.; Huang, Y.L.; Yang, W.Y.; Sampurna, B.P.; Ouchi, T.; Lee, K.L.; Wu, C.S.; Wang, H.D.; Yuh, C.H. A novel AURKA mutant-induced early-onset severe hepatocarcinogenesis greater than wild-type via activating different pathways in zebrafish. Cancers 2019, 11, 927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evason, K.J.; Francisco, M.T.; Juric, V.; Balakrishnan, S.; del Pilar Lopez Pazmino, M.; Gordan, J.D.; Kakar, S.; Spitsbergen, J.; Goga, A.; Stainier, D.Y. Identification of chemical inhibitors of β-catenin-driven liver tumorigenesis in zebrafish. PLoS Genet. 2015, 11, e1005305. [Google Scholar] [CrossRef]
- Lu, J.W.; Liao, C.Y.; Yang, W.Y.; Lin, Y.M.; Jin, S.L.; Wang, H.D.; Yuh, C.H. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS ONE 2014, 9, e85318. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Lee, J.S.; Guo, F.; Shin, J.; Perez-Atayde, A.R.; Kutok, J.L.; Rodig, S.J.; Neuberg, D.S.; Helman, D.; Feng, H.; et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 2012, 21, 362–373. [Google Scholar] [CrossRef] [Green Version]
- Modzelewska, K.; Boer, E.F.; Mosbruger, T.L.; Picard, D.; Anderson, D.; Miles, R.R.; Kroll, M.; Oslund, W.; Pysher, T.J.; Schiffman, J.D.; et al. MEK inhibitors reverse growth of embryonal brain tumors derived from oligoneural precursor cells. Cell Rep. 2016, 17, 1255–1264. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, M.W.; Liu, Y.; He, S.; Durbin, A.D.; Abraham, B.J.; Easton, J.; Shao, Y.; Xu, B.; Zhu, S.; Zhang, X.; et al. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov. 2018, 8, 320–335. [Google Scholar] [CrossRef] [Green Version]
- Gill, J.A.; Lowe, L.; Nguyen, J.; Liu, P.P.; Blake, T.; Venkatesh, B.; Aplan, P.D. Enforced expression of Simian virus 40 large T-antigen leads to testicular germ cell tumors in zebrafish. Zebrafish 2010, 7, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.E.; Bosenberg, M. Melanocytic nevi and melanoma: Unraveling a complex relationship. Oncogene 2017, 36, 5771–5792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shieh, Y.S.; Chang, Y.S.; Hong, J.R.; Chen, L.J.; Jou, L.K.; Hsu, C.C.; Her, G.M. Increase of hepatic fat accumulation by liver specific expression of Hepatitis B virus X protein in zebrafish. Biochim. Biophys. Acta 2010, 1801, 721–730. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Mansour, M.R.; Zimmerman, M.W.; Ki, D.H.; Layden, H.M.; Akahane, K.; Gjini, E.; de Groh, E.D.; Perez-Atayde, A.R.; Zhu, S.; et al. Synergy between loss of NF1 and overexpression of MYCN in neuroblastoma is mediated by the GAP-related domain. Elife 2016, 5, e14713. [Google Scholar] [CrossRef] [PubMed]
- Ignatius, M.S.; Chen, E.; Elpek, N.M.; Fuller, A.Z.; Tenente, I.M.; Clagg, R.; Liu, S.; Blackburn, J.S.; Linardic, C.M.; Rosenberg, A.E.; et al. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell 2012, 21, 680–693. [Google Scholar] [CrossRef] [Green Version]
- Sabaawy, H.E.; Azuma, M.; Embree, L.J.; Tsai, H.J.; Starost, M.F.; Hickstein, D.D. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2006, 103, 15166–15171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreasson, P.; Schwaller, J.; Anastasiadou, E.; Aster, J.; Gilliland, D.G. The expression of ETV6/CBFA2 (TEL/AML1) is not sufficient for the transformation of hematopoietic cell lines in vitro or the induction of hematologic disease in vivo. Cancer Genet. Cytogenet. 2001, 130, 93–104. [Google Scholar] [CrossRef]
- Schindler, J.W.; Van Buren, D.; Foudi, A.; Krejci, O.; Qin, J.; Orkin, S.H.; Hock, H. TEL-AML1 corrupts hematopoietic stem cells to persist in the bone marrow and initiate leukemia. Cell Stem Cell 2009, 5, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Campos-Sanchez, E.; Vicente-Dueñas, C.; Rodríguez-Hernández, G.; Capstick, M.; Kuster, N.; Dasenbrock, C.; Sánchez-García, I.; Cobaleda, C. Novel ETV6-RUNX1 mouse model to study the role of ELF-MF in childhood b-acute lymphoblastic leukemia: A pilot study. Bioelectromagnetics 2019, 40, 343–353. [Google Scholar] [CrossRef]
- Halpern, M.E.; Rhee, J.; Goll, M.G.; Akitake, C.M.; Parsons, M.; Leach, S.D. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 2008, 5, 97–110. [Google Scholar] [CrossRef]
- Fischer, J.A.; Giniger, E.; Maniatis, T.; Ptashne, M. GAL4 activates transcription in Drosophila. Nature 1988, 332, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Hartley, K.O.; Nutt, S.L.; Amaya, E. Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc. Natl. Acad. Sci. USA 2002, 99, 1377–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ornitz, D.M.; Moreadith, R.W.; Leder, P. Binary system for regulating transgene expression in mice: Targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc. Natl. Acad. Sci. USA 1991, 88, 698–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheer, N.; Campos-Ortega, J.A. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech. Dev. 1999, 80, 153–158. [Google Scholar] [CrossRef]
- Anelli, V.; Santoriello, C.; Distel, M.; Köster, R.W.; Ciccarelli, F.D.; Mione, M. Global repression of cancer gene expression in a zebrafish model of melanoma is linked to epigenetic regulation. Zebrafish 2010, 6, 417–424. [Google Scholar] [CrossRef]
- Alghisi, E.; Distel, M.; Malagola, M.; Anelli, V.; Santoriello, C.; Herwig, L.; Krudewig, A.; Henkel, C.V.; Russo, D.; Mione, M.C. Targeting oncogene expression to endothelial cells induces proliferation of the myelo-erythroid lineage by repressing the Notch pathway. Leukemia 2013, 27, 2229–2241. [Google Scholar] [CrossRef] [Green Version]
- Mayrhofer, M.; Gourain, V.; Reischl, M.; Affaticati, P.; Jenett, A.; Joly, J.S.; Benelli, M.; Demichelis, F.; Poliani, P.L.; Sieger, D.; et al. A novel brain tumour model in zebrafish reveals the role of YAP activation in MAPK- and PI3K-induced malignant growth. Dis. Model. Mech. 2017, 10, 15–28. [Google Scholar] [CrossRef] [Green Version]
- Burger, A.; Vasilyev, A.; Tomar, R.; Selig, M.K.; Nielsen, G.P.; Peterson, R.T.; Drummond, I.A.; Haber, D.A. A zebrafish model of chordoma initiated by notochord-driven expression of HRASV12. Dis. Model. Mech. 2014, 7, 907–913. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Leach, S.D. Screening pancreatic oncogenes in zebrafish using the Gal4/UAS system. Methods Cell Biol. 2011, 105, 367–381. [Google Scholar] [CrossRef] [Green Version]
- Jung, I.H.; Leem, G.L.; Jung, D.E.; Kim, M.H.; Kim, E.Y.; Kim, S.H.; Park, H.C.; Park, S.W. Glioma is formed by active Akt1 alone and promoted by active Rac1 in transgenic zebrafish. Neuro Oncol. 2013, 15, 290–304. [Google Scholar] [CrossRef] [Green Version]
- Park, J.T.; Johnson, N.; Liu, S.; Levesque, M.; Wang, Y.J.; Ho, H.; Huso, D.; Maitra, A.; Parsons, M.J.; Prescott, J.D.; et al. Differential in vivo tumorigenicity of diverse KRAS mutations in vertebrate pancreas: A comprehensive survey. Oncogene 2015, 34, 2801–2806. [Google Scholar] [CrossRef] [Green Version]
- Lakso, M.; Sauer, B.; Mosinger, B., Jr.; Lee, E.J.; Manning, R.W.; Yu, S.H.; Mulder, K.L.; Westphal, H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl. Acad. Sci. USA 1992, 89, 6232–6236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewandoski, M. Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2001, 2, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Seok, S.H.; Na, Y.R.; Han, J.H.; Kim, T.H.; Jung, H.; Lee, B.H.; Emelyanov, A.; Parinov, S.; Park, J.H. Cre/loxP-regulated transgenic zebrafish model for neural progenitor-specific oncogenic Kras expression. Cancer Sci. 2010, 101, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Park, J.T. Zebrafish model of KRAS-initiated pancreatic endocrine tumor. Anim. Cells Syst. 2019, 23, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Halloran, M.C.; Sato-Maeda, M.; Warren, J.T.; Su, F.; Lele, Z.; Krone, P.H.; Kuwada, J.Y.; Shoji, W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development 2000, 127, 1953–1960. [Google Scholar]
- Shoji, W.; Sato-Maeda, M. Application of heat shock promoter in transgenic zebrafish. Dev. Growth Differ. 2008, 50, 401–406. [Google Scholar] [CrossRef]
- Zou, J.; Guo, Y.; Guettouche, T.; Smith, D.F.; Voellmy, R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 1998, 94, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Santoriello, C.; Deflorian, G.; Pezzimenti, F.; Kawakami, K.; Lanfrancone, L.; d’Adda di Fagagna, F.; Mione, M. Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. Dis. Model. Mech. 2009, 2, 56–67. [Google Scholar] [CrossRef] [Green Version]
- Yeh, J.R.; Munson, K.M.; Chao, Y.L.; Peterson, Q.P.; Macrae, C.A.; Peterson, R.T. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development 2008, 135, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Ye, Y.; Ye, Z.; Xu, S.; Liu, W.; Xu, J.; Zhang, Y.; Liu, Q.; Huang, Z.; Zhang, W. Human BCR/ABL1 induces chronic myeloid leukemia-like disease in zebrafish. Haematologica 2020, 105, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Leacock, S.W.; Basse, A.N.; Chandler, G.L.; Kirk, A.M.; Rakheja, D.; Amatruda, J.F. A zebrafish transgenic model of Ewing’s sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesis. Dis. Model. Mech. 2012, 5, 95–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.J.; Chen, F.Y.; Zhang, Y.; Cao, L.F.; Kuang, Y.; Zhong, M.; Wang, T.; Zhong, H. MYCN transgenic zebrafish model with the characterization of acute myeloid leukemia and altered hematopoiesis. PLoS ONE 2013, 8, e59070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Zhu, J.; Chen, F.; Lin, W.; Cai, J.; Zhong, J.; Zhong, H. RUNX1-Evi-1 fusion gene inhibited differentiation and apoptosis in myelopoiesis: An in vivo study. BMC Cancer 2015, 15, 970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Langenau, D.M.; Madge, J.A.; Quinkertz, A.; Gutierrez, A.; Neuberg, D.S.; Kanki, J.P.; Look, A.T. Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br. J. Haematol. 2007, 138, 169–175. [Google Scholar] [CrossRef]
- Le, X.; Langenau, D.M.; Keefe, M.D.; Kutok, J.L.; Neuberg, D.S.; Zon, L.I. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc. Natl. Acad. Sci. USA 2007, 104, 9410–9415. [Google Scholar] [CrossRef] [Green Version]
- Forrester, A.M.; Grabher, C.; McBride, E.R.; Boyd, E.R.; Vigerstad, M.H.; Edgar, A.; Kai, F.B.; Da’as, S.I.; Payne, E.; Look, A.T.; et al. NUP98-HOXA9-transgenic zebrafish develop a myeloproliferative neoplasm and provide new insight into mechanisms of myeloid leukaemogenesis. Br. J. Haematol. 2011, 155, 167–181. [Google Scholar] [CrossRef]
- Berens, C.; Hillen, W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem. 2003, 270, 3109–3121. [Google Scholar] [CrossRef]
- Gossen, M.; Bujard, H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 1992, 89, 5547–5551. [Google Scholar] [CrossRef] [Green Version]
- Gossen, M.; Freundlieb, S.; Bender, G.; Müller, G.; Hillen, W.; Bujard, H. Transcriptional activation by tetracyclines in mammalian cells. Science 1995, 268, 1766–1769. [Google Scholar] [CrossRef]
- Stebbins, M.J.; Urlinger, S.; Byrne, G.; Bello, B.; Hillen, W.; Yin, J.C. Tetracycline-inducible systems for Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 10775–10780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furth, P.A.; St Onge, L.; Böger, H.; Gruss, P.; Gossen, M.; Kistner, A.; Bujard, H.; Hennighausen, L. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc. Natl. Acad. Sci. USA 1994, 91, 9302–9306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.J.; Jou, T.S.; Ho, Y.L.; Lee, W.H.; Jeng, Y.T.; Hsieh, F.J.; Tsai, H.J. Conditional expression of a myocardium-specific transgene in zebrafish transgenic lines. Dev. Dyn. 2005, 233, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Chen, W.; Orr, B.A.; Spitsbergen, J.M.; Jia, S.; Eden, C.J.; Henson, H.E.; Taylor, M.R. Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol. Cancer 2015, 14, 18. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Huang, X.; Zhan, H.; Zeng, Z.; Li, C.; Spitsbergen, J.M.; Meierjohann, S.; Schartl, M.; Gong, Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J. Hepatol. 2012, 56, 419–425. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, W.; Wang, Z.; Zeng, Z.; Zhan, H.; Li, C.; Zhou, L.; Yan, C.; Spitsbergen, J.M.; Gong, Z. A transgenic zebrafish liver tumor model with inducible Myc expression reveals conserved Myc signatures with mammalian liver tumors. Dis. Model. Mech. 2013, 6, 414–423. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Li, Z.; Nguyen, A.T.; Li, C.; Emelyanov, A.; Gong, Z. Xmrk, kras and myc transgenic zebrafish liver cancer models share molecular signatures with subsets of human hepatocellular carcinoma. PLoS ONE 2014, 9, e91179. [Google Scholar] [CrossRef] [Green Version]
- Chew, T.W.; Liu, X.J.; Liu, L.; Spitsbergen, J.M.; Gong, Z.; Low, B.C. Crosstalk of Ras and Rho: Activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene 2014, 33, 2717–2727. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, H.; Spitsbergen, J.M.; Gong, Z. Males develop faster and more severe hepatocellular carcinoma than females in kras(V12) transgenic zebrafish. Sci. Rep. 2017, 7, 41280. [Google Scholar] [CrossRef]
- Yang, Q.; Yan, C.; Wang, X.; Gong, Z. Leptin induces muscle wasting in a zebrafish kras-driven hepatocellular carcinoma (HCC) model. Dis. Model. Mech. 2019, 12, dmm038240. [Google Scholar] [CrossRef] [Green Version]
- Evans, R.M. The steroid and thyroid hormone receptor superfamily. Science 1988, 240, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.J.; Green, S.; Jin, J.R.; Chambon, P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell 1988, 54, 199–207. [Google Scholar] [CrossRef]
- Picard, D.; Salser, S.J.; Yamamoto, K.R. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell 1988, 54, 1073–1080. [Google Scholar] [CrossRef]
- Eilers, M.; Picard, D.; Yamamoto, K.R.; Bishop, J.M. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature 1989, 340, 66–68. [Google Scholar] [CrossRef]
- Jackson, P.; Baltimore, D.; Picard, D. Hormone-conditional transformation by fusion proteins of c-Abl and its transforming variants. Embo J. 1993, 12, 2809–2819. [Google Scholar] [CrossRef]
- Picard, D. Regulation of protein function through expression of chimaeric proteins. Curr. Opin. Biotechnol. 1994, 5, 511–515. [Google Scholar] [CrossRef]
- Gutierrez, A.; Grebliunaite, R.; Feng, H.; Kozakewich, E.; Zhu, S.; Guo, F.; Payne, E.; Mansour, M.; Dahlberg, S.E.; Neuberg, D.S.; et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J. Exp. Med. 2011, 208, 1595–1603. [Google Scholar] [CrossRef]
- Littlewood, T.D.; Hancock, D.C.; Danielian, P.S.; Parker, M.G.; Evan, G.I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995, 23, 1686–1690. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, J.; Lu, J.W.; Makinoshima, H.; Gong, Z. A novel zebrafish model of metastasis identifies the hsd11β1 inhibitor adrenosterone as a suppressor of epithelial-mesenchymal transition and metastatic dissemination. Mol. Cancer Res. 2020, 18, 477–487. [Google Scholar] [CrossRef]
- Baulieu, E.E. Contragestion and other clinical applications of RU 486, an antiprogesterone at the receptor. Science 1989, 245, 1351–1357. [Google Scholar] [CrossRef]
- Wang, Y.; O’Malley, B.W., Jr.; Tsai, S.Y.; O’Malley, B.W. A regulatory system for use in gene transfer. Proc. Natl. Acad. Sci. USA 1994, 91, 8180–8184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellendonk, C.; Tronche, F.; Monaghan, A.P.; Angrand, P.O.; Stewart, F.; Schütz, G. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res. 1996, 24, 1404–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emelyanov, A.; Parinov, S. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 2008, 320, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.; Spitsbergen, J.M.; Parinov, S.; Gong, Z. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis. Model. Mech. 2012, 5, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Yang, Q.; Shen, H.M.; Spitsbergen, J.M.; Gong, Z. Chronically high level of tgfb1a induction causes both hepatocellular carcinoma and cholangiocarcinoma via a dominant Erk pathway in zebrafish. Oncotarget 2017, 8, 77096–77109. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Yangn, Q.; Gong, Z. Transgenic expression of tgfb1a induces hepatic inflammation, fibrosis and metastasis in zebrafish. Biochem. Biophys. Res. Commun. 2019, 509, 175–181. [Google Scholar] [CrossRef]
- Kenyon, A.; Gavriouchkina, D.; Zorman, J.; Chong-Morrison, V.; Napolitani, G.; Cerundolo, V.; Sauka-Spengler, T. Generation of a double binary transgenic zebrafish model to study myeloid gene regulation in response to oncogene activation in melanocytes. Dis. Model. Mech. 2018, 11, dmm030056. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.W.; Raghuram, D.; Fong, P.A.; Gong, Z. Inducible intestine-specific expression of krasv12 triggers intestinal tumorigenesis in transgenic zebrafish. Neoplasia 2018, 20, 1187–1197. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Koh, V.; Spitsbergen, J.M.; Gong, Z. Development of a conditional liver tumor model by mifepristone-inducible Cre recombination to control oncogenic kras V12 expression in transgenic zebrafish. Sci. Rep. 2016, 6, 19559. [Google Scholar] [CrossRef] [Green Version]
- Schwenk, F.; Kuhn, R.; Angrand, P.O.; Rajewsky, K.; Stewart, A.F. Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res. 1998, 26, 1427–1432. [Google Scholar] [CrossRef]
- Indra, A.K.; Warot, X.; Brocard, J.; Bornert, J.M.; Xiao, J.H.; Chambon, P.; Metzger, D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999, 27, 4324–4327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jopling, C.; Sleep, E.; Raya, M.; Martí, M.; Raya, A.; Izpisúa Belmonte, J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Jungke, P.; Hans, S.; Brand, M. The zebrafish CreZoo: An easy-to-handle database for novel CreER(T2)-driver lines. Zebrafish 2013, 10, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Kalasekar, S.M.; Kotiyal, S.; Conley, C.; Phan, C.; Young, A.; Evason, K.J. Heterogeneous beta-catenin activation is sufficient to cause hepatocellular carcinoma in zebrafish. Biol. Open 2019, 8, bio047829. [Google Scholar] [CrossRef] [Green Version]
- Park, J.T.; Leach, S.D. Zebrafish model of KRAS-initiated pancreatic cancer. Anim. Cells Syst. 2018, 22, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Tomasetti, C.; Marchionni, L.; Nowak, M.A.; Parmigiani, G.; Vogelstein, B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc. Natl. Acad. Sci. USA 2015, 112, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Hornsby, C.; Page, K.M.; Tomlinson, I.P. What can we learn from the population incidence of cancer? Armitage and Doll revisited. Lancet Oncol. 2007, 8, 1030–1038. [Google Scholar] [CrossRef]
- Häusser, M. Optogenetics: The age of light. Nat. Methods 2014, 11, 1012–1014. [Google Scholar] [CrossRef]
- Zhang, K.; Cui, B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 2015, 33, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Rogers, K.W.; Müller, P. Optogenetic approaches to investigate spatiotemporal signaling during development. Curr. Top. Dev. Biol. 2020, 137, 37–77. [Google Scholar] [CrossRef]
- Feng, Z.; Nam, S.; Hamouri, F.; Aujard, I.; Ducos, B.; Vriz, S.; Volovitch, M.; Jullien, L.; Lin, S.; Weiss, S.; et al. Optical control of tumor induction in the zebrafish. Sci. Rep. 2017, 7, 9195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ung, C.Y.; Guo, F.; Zhang, X.; Zhu, Z.; Zhu, S. Mosaic zebrafish transgenesis for functional genomic analysis of candidate cooperative genes in tumor pathogenesis. J. Vis. Exp. 2015, 52567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köster, R.W.; Fraser, S.E. Tracing transgene expression in living zebrafish embryos. Dev. Biol. 2001, 233, 329–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langenau, D.M.; Keefe, M.D.; Storer, N.Y.; Jette, C.A.; Smith, A.C.; Ceol, C.J.; Bourque, C.; Look, A.T.; Zon, L.I. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene 2008, 27, 4242–4248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceol, C.J.; Houvras, Y.; Jane-Valbuena, J.; Bilodeau, S.; Orlando, D.A.; Battisti, V.; Fritsch, L.; Lin, W.M.; Hollmann, T.J.; Ferré, F.; et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011, 471, 513–517. [Google Scholar] [CrossRef]
- McConnell, A.M.; Mito, J.K.; Ablain, J.; Dang, M.; Formichella, L.; Fisher, D.E.; Zon, L.I. Neural crest state activation in NRAS driven melanoma, but not in NRAS-driven melanocyte expansion. Dev. Biol. 2019, 449, 107–114. [Google Scholar] [CrossRef]
- Hosono, Y.; Niknafs, Y.S.; Prensner, J.R.; Iyer, M.K.; Dhanasekaran, S.M.; Mehra, R.; Pitchiaya, S.; Tien, J.; Escara-Wilke, J.; Poliakov, A.; et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 2017, 171, 1559–1572. [Google Scholar] [CrossRef]
- Callahan, S.J.; Tepan, S.; Zhang, Y.M.; Lindsay, H.; Burger, A.; Campbell, N.R.; Kim, I.S.; Hollmann, T.J.; Studer, L.; Mosimann, C.; et al. Cancer modeling by transgene electroporation in adult zebrafish (TEAZ). Dis. Model. Mech. 2018, 11, dmm034561. [Google Scholar] [CrossRef] [Green Version]
- Ung, C.Y.; Lam, S.H.; Gong, Z. Comparative transcriptome analyses revealed conserved biological and transcription factor target modules between the zebrafish and human tumors. Zebrafish 2009, 6, 425–431. [Google Scholar] [CrossRef]
- Zhang, G.; Hoersch, S.; Amsterdam, A.; Whittaker, C.A.; Beert, E.; Catchen, J.M.; Farrington, S.; Postlethwait, J.H.; Legius, E.; Hopkins, N.; et al. Comparative oncogenomic analysis of copy number alterations in human and zebrafish tumors enables cancer driver discovery. PLoS Genet. 2013, 9, e1003734. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, J.; Makinoshima, H. Zebrafish-based screening models for the identification of anti-metastatic drugs. Molecules 2020, 25, 2407. [Google Scholar] [CrossRef] [PubMed]
- Fazio, M.; Ablain, J.; Chuan, Y.; Langenau, D.M.; Zon, L.I. Zebrafish patient avatars in cancer biology and precision cancer therapy. Nat. Rev. Cancer 2020, 20, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jette, C.; Kanki, J.P.; Aster, J.C.; Look, A.T.; Griffin, J.D. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia 2007, 21, 462–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liau, W.S.; Tan, S.H.; Ngoc, P.C.T.; Wang, C.Q.; Tergaonkar, V.; Feng, H.; Gong, Z.; Osato, M.; Look, A.T.; Sanda, T. Aberrant activation of the GIMAP enhancer by oncogenic transcription factors in T-cell acute lymphoblastic leukemia. Leukemia 2017, 31, 1798–1807. [Google Scholar] [CrossRef]
- Langenau, D.M.; Jette, C.; Berghmans, S.; Palomero, T.; Kanki, J.P.; Kutok, J.L.; Look, A.T. Suppression of apoptosis by bcl-2 overexpression in lymphoid cells of transgenic zebrafish. Blood 2005, 105, 3278–3285. [Google Scholar] [CrossRef] [Green Version]
- Onnebo, S.M.; Condron, M.M.; McPhee, D.O.; Lieschke, G.J.; Ward, A.C. Hematopoietic perturbation in zebrafish expressing a tel-jak2a fusion. Exp. Hematol. 2005, 33, 182–188. [Google Scholar] [CrossRef]
- Onnebo, S.M.; Rasighaemi, P.; Kumar, J.; Liongue, C.; Ward, A.C. Alternative TEL-JAK2 fusions associated with T-cell acute lymphoblastic leukemia and atypical chronic myelogenous leukemia dissected in zebrafish. Haematologica 2012, 97, 1895–1903. [Google Scholar] [CrossRef] [Green Version]
- Lister, J.A.; Capper, A.; Zeng, Z.; Mathers, M.E.; Richardson, J.; Paranthaman, K.; Jackson, I.J.; Patton, E.E. A conditional zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion vs. regression in vivo. J. Invest. Derm. 2014, 134, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Santoriello, C.; Gennaro, E.; Anelli, V.; Distel, M.; Kelly, A.; Köster, R.W.; Hurlstone, A.; Mione, M. Kita driven expression of oncogenic HRAS leads to early onset and highly penetrant melanoma in zebrafish. PLoS ONE 2010, 5, e15170. [Google Scholar] [CrossRef] [Green Version]
- Iwanaga, R.; Truong, B.T.; Lambert, K.A.; Vyas, R.; Orlicky, D.; Shellman, Y.G.; Tan, A.C.; Artinger, K.B. Loss of prdm1a accelerates melanoma onset and progression. bioRxiv 2019. Epub ahead of print. PMID: 32562448. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.Y.; Chen, C.F.; Rajendran, R.S.; Shen, C.N.; Chen, T.H.; Yen, C.C.; Chuang, C.K.; Lin, D.S.; Hsiao, C.D. Overexpression of Akt1 enhances adipogenesis and leads to lipoma formation in zebrafish. PLoS ONE 2012, 7, e36474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, B.; Chen, W.; Spitsbergen, J.M.; Lu, J.; Vogel, P.; Peters, J.L.; Wang, Y.D.; Orr, B.A.; Wu, J.; Henson, H.E.; et al. Activation of sonic hedgehog signaling in neural progenitor cells promotes glioma development in the zebrafish optic pathway. Oncogenesis 2014, 3, e96. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhang, X.; Weichert-Leahey, N.; Dong, Z.; Zhang, C.; Lopez, G.; Tao, T.; He, S.; Wood, A.C.; Oldridge, D.; et al. LMO1 synergizes with MYCN to promote neuroblastoma initiation and metastasis. Cancer Cell 2017, 32, C310–C323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, T.; Shi, H.; Mariani, L.; Abraham, B.J.; Durbin, A.D.; Zimmerman, M.W.; Powers, J.T.; Missios, P.; Ross, K.N.; Perez-Atayde, A.R.; et al. LIN28B regulates transcription and potentiates MYCN-induced neuroblastoma through binding to ZNF143 at target gene promotors. Proc. Natl. Acad. Sci. USA 2020, 28, 16516–16526. [Google Scholar] [CrossRef] [PubMed]
- Balci, T.B.; Prykhozhij, S.V.; The, E.M.; Da’as, S.I.; McBride, E.; Liwski, R.; Chute, I.C.; Leger, D.; Lewis, S.M.; Berman, J.N. A transgenic zebrafish model expressing KIT-D816V recapitulates features of aggressive systemic mastocytosis. Br. J. Haematol. 2014, 167, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.A.; Jiang, H.; Ben-Shlomo, A.; Wawrowsky, K.; Fan, X.M.; Lin, S.; Melmed, S. Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin-dependent kinase (CDK) inhibitor. Proc. Natl. Acad. Sci. USA 2011, 108, 8414–8419. [Google Scholar] [CrossRef] [Green Version]





| Chemicals | Cancer Types | References |
|---|---|---|
| NDMA | Hepatocarinoma | [26] |
| DEN | Liver and pancreas carcinomas | [27] |
| DMBA | Liver, gill, pancreas, gastrointestinal, pancreas, testis, cartilage, muscle, blood vessel, connective and lymphoid tissues and neural tumors | [28,29] |
| DBP | Hepatocarcinoma | [29] |
| MNNG | Liver, gill, pancreas, gastrointestinal, pancreas, testis, kidney, muscle tumors and hemangio(sarco)mas | [30] |
| ENU | Epidermal papillomas | [31] |
| Mutagenesis | Mutated Genes | Cancer Phenotypes | References |
|---|---|---|---|
| ENU | dnaaf1 (lrcc50) | Seminoma (incidence: 90% after 2 years of age) | [40] |
| ENU | bmpr1bb (alk6b) | Testicular germ cell tumor | [41] |
| Retroviral insertion | Ribosomal proteins (rps8, rps15a, rpl7, rpl35, rpl36, rpl36a, rpl13, rpl23a, rps7, rps18, rps29) | MPNST (incidence: 14% to 100% at 2 years of age) | [43] |
| Retroviral insertion | nf2a | MPNST | [43] |
| Retroviral insertion | fgf8 (overexpression) | Neuroblastoma (incidence: 25–50% at 2 years of age) | [44] |
| ENU | mybl2b (bmyb) (Mutant crash&burn) | Altered cell proliferation and genome instability Increased MNNG-induced cancer susceptibility | [45] |
| ENU | espl1 (separase) (Mutant cease&desist) | High levels of polyploidy and aneuploidy. Increased MNNG-induced cancer susceptibility | [46] |
| ENU | sufu (Mutant dre) hhip (Mutant uki) ptch1 (Mutant lep) | Activation of the Hedgehog signaling pathway | [47] |
| ENU | tp53 | Deficiency in radiation-induced apoptosis Sarcomas (100% penetrance in adults) | [50] |
| ENU | llgl2 (lgl2) (Mutant pen) | Epidermal cell tumor | [51,52] |
| ENU | myh11 (Mutant mlt) | Epithelial invasion and cystic expansion of the intestine | [53] |
| Gene | Cancer Types | References |
|---|---|---|
| tp53 | MPNSTs (Incidence: 28% at 8.5 months of age) | [60] |
| ptena, ptenb | Ocular hemangiosarcomas (Incidence: 10.2% at 12 months of age) | [61,62] |
| apc | Intestinal, hepatic and pancreatic proliferative neoplasia (Incidence: 29% at 15 months of age) | [63] |
| mlh1, msh2, msh6 | Primarily neurofibromas in the eye and abdomen. (Incidence: about 33% at 18 months of age) | [64] |
| brca2 | Testicular Neoplasia (Incidence: 31% at 10–16 months of age) | [65] |
| Nuclease | Mutated Genes | Cancer Types | References |
|---|---|---|---|
| ZFN | nf1a, nf1b | High grade gliomas and MPNSTs in the tp53−/−genetic background | [76] |
| ZFN | tet2 | Generalized MDS at 24 months of age | [77] |
| TALEN | rb1 | Primitive neuroectodermal tumors (Incidence: 11–33% at 3.5 months of age in F0 mosaics) | [78,79,80] |
| TALEN | cdkn2a/b | MPNSTs in F0 tp53−/−mutants (Incidence: 39% at 35 weeks of age) | [79] |
| TALEN | tp53del/del | Angiosarcoma, MPNSTs, germ cell tumors (Incidence: 37% at 12 months of age) | [81] |
| TALEN CRISPR | irx1a irx1b | Intestinal hyperplasia and testicular, ovarian, renal and bile duct tumors | [82] |
| CRISPR | spred1, tp53, ptena/b, cdkn2a | Melanocyte-specific tumor suppressor gene inactivation promotes KIT-, BRAF- and NRAS-driven melanoma formation | [83] |
| CRISPR | ptch1 | T-cell-specific inactivation promotes NOTCH1ICD-driven T-ALL development | [84] |
| CRISPR | atrx | Hemizygous loss of atrx promotes the development of epithelioid sarcoma, angiosarcoma and pleomorphic sarcoma in tp53/nf1-deficient mutants | [85] |
| CRISPR | suz12a, suz12b | Loss of suz12 function accelerates the onset of MPNST and expands the spectrum of tumor types in tp53/nf1-deficient mutants | [86] |
| CRISPR | arid1aa, arid1ab | arid1aa or arid1ab deficiency increases the penetrance of MYCN-induced neuroblastoma in Tg(dbh:EGFP-MYCN) transgenics | [87] |
| CRISPR | lats2 | PNST in F0 mosaics | [88] |
| CRISPR | dact2 | Tumor formation in the digestive system (active proliferation and hyperplasia in the pancreatic duct, the intrahepatic bile duct, the intestinal epithelium, pancreas and liver) | [89] |
| CRISPR | twist3 | Loss of twist3 prevents BRAFV600E-mediated tumorigenesis in thyroid | [90] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raby, L.; Völkel, P.; Le Bourhis, X.; Angrand, P.-O. Genetic Engineering of Zebrafish in Cancer Research. Cancers 2020, 12, 2168. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12082168
Raby L, Völkel P, Le Bourhis X, Angrand P-O. Genetic Engineering of Zebrafish in Cancer Research. Cancers. 2020; 12(8):2168. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12082168
Chicago/Turabian StyleRaby, Ludivine, Pamela Völkel, Xuefen Le Bourhis, and Pierre-Olivier Angrand. 2020. "Genetic Engineering of Zebrafish in Cancer Research" Cancers 12, no. 8: 2168. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12082168