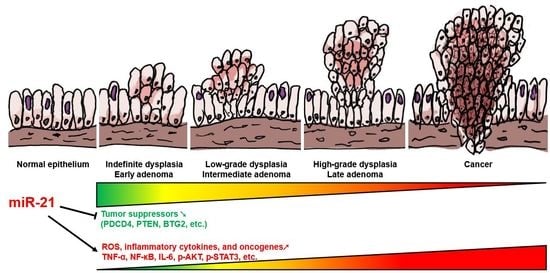
MicroRNA-21 Plays Multiple Oncometabolic Roles in Colitis-Associated Carcinoma and Colorectal Cancer via the PI3K/AKT, STAT3, and PDCD4/TNF-α Signaling Pathways in Zebrafish
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Zebrafish Lines and Maintenance
2.3. Doxycycline (Dox) Treatment
2.4. Chemical Treatment
2.5. RNA Analysis
2.6. Whole-Mount Alcian Blue Staining
2.7. Histopathology
2.8. Western Blotting Analysis
2.9. Immunohistochemistry
2.10. Statistical Analysis
3. Results
3.1. Generation of the Transgenic Zebrafish Line ImiR-21
3.2. Identification of Intestinal Inflammation Markers in Zebrafish
3.3. Effects of Intestinal miR-21 Expression on Early Onset of IBD-Like Colitis
3.4. Overexpression of miR-21 Causes Intestinal Epithelial Barrier Impairment
3.5. Chronic Effects of Intestinal miR-21 Expression on Colitis
3.6. Chronic Effects of Intestinal miR-21 Expression on CAC
3.7. miR21 Promotes CAC Development by Activating PI3K/AKT, IL-6/JAK/STAT3, and PDCD4/NF-κB/TNF-α (PSP) Signaling Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.E.M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Available online: https://gco.iarc.fr/today (accessed on 1 July 2021).
- American Cancer Society. What is Colorectal Cancer? Available online: https://www.cancer.org/cancer/colon-rectal-cancer/about/what-is-colorectal-cancer.html (accessed on 1 July 2021).
- Kulaylat, M.N.; Dayton, M.T. Ulcerative colitis and cancer. J. Surg. Oncol. 2010, 101, 706–712. [Google Scholar] [CrossRef]
- Fiocchi, C. Inflammatory bowel disease: Etiology and pathogenesis. Gastroenterology 1998, 115, 182–205. [Google Scholar] [CrossRef]
- Beaugerie, L.; Itzkowitz, S.H. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 2015, 372, 1441–1452. [Google Scholar] [CrossRef]
- Rutter, M.; Saunders, B.; Wilkinson, K.; Rumbles, S.; Schofield, G.; Kamm, M.; Williams, C.; Price, A.; Talbot, I.; Forbes, A. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology 2004, 126, 451–459. [Google Scholar] [CrossRef]
- Grivennikov, S.I. Inflammation and colorectal cancer: Colitis-associated neoplasia. Semin. Immunopathol. 2013, 35, 229–244. [Google Scholar] [CrossRef]
- Chiba, T.; Marusawa, H.; Ushijima, T. Inflammation-associated cancer development in digestive organs: Mechanisms and roles for genetic and epigenetic modulation. Gastroenterology 2012, 143, 550–563. [Google Scholar] [CrossRef] [Green Version]
- Hyun, Y.S.; Han, D.S.; Lee, A.R.; Eun, C.S.; Youn, J.; Kim, H.Y. Role of il-17a in the development of colitis-associated cancer. Carcinogenesis 2012, 33, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Popivanova, B.K.; Kitamura, K.; Wu, Y.; Kondo, T.; Kagaya, T.; Kaneko, S.; Oshima, M.; Fujii, C.; Mukaida, N. Blocking tnf-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J. Clin. Investig. 2008, 118, 560–570. [Google Scholar]
- Wang, X.; Yang, L.; Huang, F.; Zhang, Q.; Liu, S.; Ma, L.; You, Z. Inflammatory cytokines il-17 and tnf-alpha up-regulate pd-l1 expression in human prostate and colon cancer cells. Immunol. Lett. 2017, 184, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Al-Ismaeel, Q.; Neal, C.P.; Al-Mahmoodi, H.; Almutairi, Z.; Al-Shamarti, I.; Straatman, K.; Jaunbocus, N.; Irvine, A.; Issa, E.; Moreman, C.; et al. Zeb1 and il-6/11-stat3 signalling cooperate to define invasive potential of pancreatic cancer cells via differential regulation of the expression of s100 proteins. Br. J. Cancer 2019, 121, 65–75. [Google Scholar] [CrossRef]
- Heichler, C.; Scheibe, K.; Schmied, A.; Geppert, C.I.; Schmid, B.; Wirtz, S.; Thoma, O.M.; Kramer, V.; Waldner, M.J.; Buttner, C.; et al. Stat3 activation through il-6/il-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut 2020, 69, 1269–1282. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting stat3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Hirano, T. Il-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef]
- Ma, J.H.; Qin, L.; Li, X. Role of stat3 signaling pathway in breast cancer. Cell Commun. Signal 2020, 18, 33. [Google Scholar] [CrossRef] [Green Version]
- Bollrath, J.; Phesse, T.J.; von Burstin, V.A.; Putoczki, T.; Bennecke, M.; Bateman, T.; Nebelsiek, T.; Lundgren-May, T.; Canli, O.; Schwitalla, S.; et al. Gp130-mediated stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 2009, 15, 91–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. Il-6 and stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Waldner, M.J.; Neurath, M.F. Master regulator of intestinal disease: Il-6 in chronic inflammation and cancer development. Semin. Immunol. 2014, 26, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, M.; Guo, C.; Wang, G.; Zhu, F.; Wang, J.; Wang, X.; Wang, Q.; Zhao, W.; Shi, Y.; et al. Pdcd4 deficiency aggravated colitis and colitis-associated colorectal cancer via promoting il-6/stat3 pathway in mice. Inflamm. Bowel Dis. 2016, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.; Yin, Y.; Guo, H.; Li, S.; Sun, Y.; Zeng, C.; Zhu, W. The mechanisms and clinical significance of pdcd4 in colorectal cancer. Gene 2019, 680, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Lankat-Buttgereit, B.; Goke, R. The tumour suppressor pdcd4: Recent advances in the elucidation of function and regulation. Biol. Cell 2009, 101, 309–317. [Google Scholar] [CrossRef]
- Wang, Q.; Dong, Z.; Liu, X.; Song, X.; Song, Q.; Shang, Q.; Jiang, Y.; Guo, C.; Zhang, L. Programmed cell death-4 deficiency prevents diet-induced obesity, adipose tissue inflammation, and insulin resistance. Diabetes 2013, 62, 4132–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A. Negative regulation of tlr4 via targeting of the proinflammatory tumor suppressor pdcd4 by the microrna mir-21. Nat. Immunol. 2010, 11, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Wang, T.; Kameswaran, V.; Wei, Q.; Johnson, D.S.; Matschinsky, F.; Shi, W.; Chen, Y.H. The microrna-21-pdcd4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc. Natl. Acad. Sci. USA 2011, 108, 12030–12035. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Yu, Y.; Tan, S. The role of the mir-21-5p-mediated inflammatory pathway in ulcerative colitis. Exp. Ther. Med. 2020, 19, 981–989. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Zhang, X.; Xu, Y. Aberrant expression of mir-21 in patients with inflammatory bowel disease: A protocol for systematic review and meta analysis. Medicine 2020, 99, e19693. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. Microrna-21 (mir-21) post-transcriptionally downregulates tumor suppressor pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef] [Green Version]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef]
- Hilton, I.B.; Gersbach, C.A. Enabling functional genomics with genome engineering. Genome Res. 2015, 25, 1442–1455. [Google Scholar] [CrossRef] [Green Version]
- Kobar, K.; Collett, K.; Prykhozhij, S.V.; Berman, J.N. Zebrafish cancer predisposition models. Front. Cell Dev. Biol 2021, 9, 660069. [Google Scholar] [CrossRef]
- Casey, M.J.; Stewart, R.A. Pediatric cancer models in zebrafish. Trends Cancer 2020, 6, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Stoletov, K.; Klemke, R. Catch of the day: Zebrafish as a human cancer model. Oncogene 2008, 27, 4509–4520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feitsma, H.; Cuppen, E. Zebrafish as a cancer model. Mol. Cancer Res. 2008, 6, 685–694. [Google Scholar] [CrossRef] [Green Version]
- White, R.M. Genomic approaches to zebrafish cancer. Adv. Exp. Med. Biol. 2016, 916, 125–145. [Google Scholar]
- Zhao, X.; Pack, M. Modeling intestinal disorders using zebrafish. Methods Cell Biol. 2017, 138, 241–270. [Google Scholar] [PubMed]
- Neal, J.T.; Peterson, T.S.; Kent, M.L.; Guillemin, K.H. Pylori virulence factor caga increases intestinal cell proliferation by wnt pathway activation in a transgenic zebrafish model. Dis. Model Mech. 2013, 6, 802–810. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.W.; Raghuram, D.; Fong, P.A.; Gong, Z. Inducible intestine-specific expression of kras(v12) triggers intestinal tumorigenesis in transgenic zebrafish. Neoplasia 2018, 20, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Sun, Y.; Fong, P.A.; Lin, L.I.; Liu, D.; Gong, Z. Lipopolysaccharides enhance epithelial hyperplasia and tubular adenoma in intestine-specific expression of kras(v)(12) in transgenic zebrafish. Biomedicines 2021, 9, 974. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Gao, S.; Zhang, B.; Shi, W.; Li, A.; Tian, Q. The critical role of the mir-21-meg2 axis in colorectal cancer. Crit. Rev. Eukaryot. Gene Expr. 2020, 30, 509–518. [Google Scholar] [CrossRef]
- Liu, T.; Liu, D.; Guan, S.; Dong, M. Diagnostic role of circulating mir-21 in colorectal cancer: A update meta-analysis. Ann. Med. 2021, 53, 87–102. [Google Scholar] [CrossRef]
- Lai, C.Y.; Yeh, K.Y.; Lin, C.Y.; Hsieh, Y.W.; Lai, H.H.; Chen, J.R.; Hsu, C.C.; Her, G.M. Microrna-21 plays multiple oncometabolic roles in the process of nafld-related hepatocellular carcinoma via pi3k/akt, tgf-beta, and stat3 signaling. Cancers 2021, 13, 940. [Google Scholar] [CrossRef]
- Dong, M.; Fu, Y.F.; Du, T.T.; Jing, C.B.; Fu, C.T.; Chen, Y.; Jin, Y.; Deng, M.; Liu, T.X. Heritable and lineage-specific gene knockdown in zebrafish embryo. PLoS ONE 2009, 4, e6125. [Google Scholar] [CrossRef] [Green Version]
- Sheedy, F.J. Turning 21: Induction of mir-21 as a key switch in the inflammatory response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wan, X.; Ruan, Q. The microrna-21 in autoimmune diseases. Int. J. Mol. Sci. 2016, 17, 864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, J.; Huang, Q.; Wang, L.; Ma, X.; Deng, Q.; Kumar, M.; Zhou, Z.; Li, L.; Zeng, Z.; Young, K.H.; et al. Mir-21 depletion in macrophages promotes tumoricidal polarization and enhances pd-1 immunotherapy. Oncogene 2018, 37, 3151–3165. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Sun, Y.; Wang, D.; Zheng, S.; Zhang, J.; Zheng, C. Celastrol ameliorates ulcerative colitis-related colorectal cancer in mice via suppressing inflammatory responses and epithelial-mesenchymal transition. Front. Pharmacol. 2015, 6, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Constante, M.; Fragoso, G.; Calve, A.; Samba-Mondonga, M.; Santos, M.M. Dietary heme induces gut dysbiosis, aggravates colitis, and potentiates the development of adenomas in mice. Front. Microbiol. 2017, 8, 1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacouton, E.; Chain, F.; Sokol, H.; Langella, P.; Bermudez-Humaran, L.G. Probiotic strain lactobacillus casei bl23 prevents colitis-associated colorectal cancer. Front. Immunol. 2017, 8, 1553. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, L.; Wang, J.; Qin, Z.; Wang, J.; Lu, Y.; Zheng, X.; Peng, Q.; Ye, Q.; Ai, F.; et al. Suppression colitis and colitis-associated colon cancer by anti-s100a9 antibody in mice. Front. Immunol. 2017, 8, 1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Ma, Y.; Shi, C.; Chen, H.; Zhang, H.; Chen, N.; Zhang, P.; Wang, F.; Yang, J.; Yang, J.; et al. Overexpression of mir-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the rho gtpase rhob. Biochem Biophys Res. Commun 2013, 434, 746–752. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Novellasdemunt, L.; Antas, P.; Li, V.S. Targeting wnt signaling in colorectal cancer. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am. J. Physiol. Cell Physiol. 2015, 309, C511–C521. [Google Scholar] [CrossRef]
- Oguma, K.; Oshima, H.; Aoki, M.; Uchio, R.; Naka, K.; Nakamura, S.; Hirao, A.; Saya, H.; Taketo, M.M.; Oshima, M. Activated macrophages promote wnt signalling through tumour necrosis factor-alpha in gastric tumour cells. EMBO J. 2008, 27, 1671–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greten, F.R.; Grivennikov, S.I. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Kaler, P.; Godasi, B.N.; Augenlicht, L.; Klampfer, L. The nf-kappab/akt-dependent induction of wnt signaling in colon cancer cells by macrophages and il-1beta. Cancer Microenviron. 2009, 2, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaler, P.; Augenlicht, L.; Klampfer, L. Macrophage-derived il-1beta stimulates wnt signaling and growth of colon cancer cells: A crosstalk interrupted by vitamin d3. Oncogene 2009, 28, 3892–3902. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, L.; Wei, Z.; Zhang, X.; Gao, Q.; Ma, Y.; Liu, X.; Jiang, Y.; Liu, X.; Guo, C.; et al. The inhibitory action of pdcd4 in lipopolysaccharide/d-galactosamine-induced acute liver injury. Lab. Invest. 2013, 93, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Hilliard, A.; Hilliard, B.; Zheng, S.J.; Sun, H.; Miwa, T.; Song, W.; Goke, R.; Chen, Y.H. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J. Immunol. 2006, 177, 8095–8102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Zhang, Z. Tnf-alpha promotes colon cancer cell migration and invasion by upregulating trop-2. Oncol. Lett. 2018, 15, 3820–3827. [Google Scholar]
- Liu, R.Y.; Zeng, Y.; Lei, Z.; Wang, L.; Yang, H.; Liu, Z.; Zhao, J.; Zhang, H.T. Jak/stat3 signaling is required for tgf-beta-induced epithelial-mesenchymal transition in lung cancer cells. Int. J. Oncol. 2014, 44, 1643–1651. [Google Scholar] [CrossRef] [Green Version]
- Kanehara, K.; Ohnuma, S.; Kanazawa, Y.; Sato, K.; Kokubo, S.; Suzuki, H.; Karasawa, H.; Suzuki, T.; Suzuki, C.; Naitoh, T.; et al. The indole compound ma-35 attenuates tumorigenesis in an inflammation-induced colon cancer model. Sci. Rep. 2019, 9, 12739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, C.W.; Fingleton, B.; Williams, A.; Ning, W.; Fischer, M.A.; Washington, M.K.; Chaturvedi, R.; Wilson, K.T.; Hiebert, S.W.; Williams, C.S. Mtgr1 is required for tumorigenesis in the murine aom/dss colitis-associated carcinoma model. Cancer Res. 2011, 71, 1302–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum mir-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef] [Green Version]
- Dehghan, F.; Boozarpour, S.; Torabizadeh, Z.; Alijanpour, S. Mir-21: A promising biomarker for the early detection of colon cancer. OncoTargets Ther. 2019, 12, 5601–5607. [Google Scholar] [CrossRef] [PubMed] [Green Version]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, C.-Y.; Yeh, K.-Y.; Liu, B.-F.; Chang, T.-M.; Chang, C.-H.; Liao, Y.-F.; Liu, Y.-W.; Her, G.M. MicroRNA-21 Plays Multiple Oncometabolic Roles in Colitis-Associated Carcinoma and Colorectal Cancer via the PI3K/AKT, STAT3, and PDCD4/TNF-α Signaling Pathways in Zebrafish. Cancers 2021, 13, 5565. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13215565
Lai C-Y, Yeh K-Y, Liu B-F, Chang T-M, Chang C-H, Liao Y-F, Liu Y-W, Her GM. MicroRNA-21 Plays Multiple Oncometabolic Roles in Colitis-Associated Carcinoma and Colorectal Cancer via the PI3K/AKT, STAT3, and PDCD4/TNF-α Signaling Pathways in Zebrafish. Cancers. 2021; 13(21):5565. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13215565
Chicago/Turabian StyleLai, Chi-Yu, Kun-Yun Yeh, Bi-Feng Liu, Tzu-Ming Chang, Chuan-Hsun Chang, Yung-Feng Liao, Yi-Wen Liu, and Guor Mour Her. 2021. "MicroRNA-21 Plays Multiple Oncometabolic Roles in Colitis-Associated Carcinoma and Colorectal Cancer via the PI3K/AKT, STAT3, and PDCD4/TNF-α Signaling Pathways in Zebrafish" Cancers 13, no. 21: 5565. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13215565