Multiparametric Phenotyping of Circulating Tumor Cells for Analysis of Therapeutic Targets, Oncogenic Signaling Pathways and DNA Repair Markers
Abstract
:Simple Summary
Abstract
1. Introduction
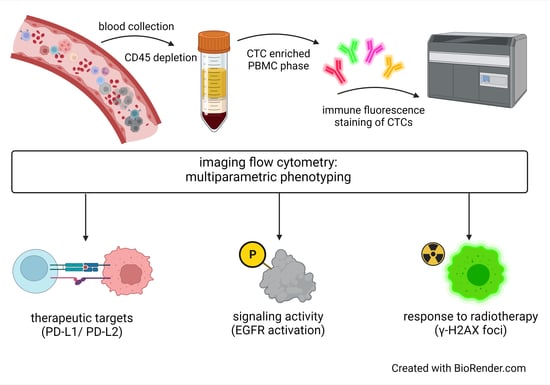
2. Materials and Methods
2.1. Cell Lines
2.2. Blood Collection
2.3. Blood Sample Processing for CTC Enrichment
2.4. Immune Fluorescence (IF) Staining
2.5. Imaging Flow Cytometry—Amnis® ImageStream®X Mk II
2.6. Spiking Experiments
3. Results
3.1. Establishment of Multicolor CTC Detection
3.2. Phenotyping of CTCs
3.3. Assessment of CTCs in Blood Samples from Patients with HNSCC and BC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lozar, T.; Gersak, K.; Cemazar, M.; Kuhar, C.G.; Jesenko, T. The biology and clinical potential of circulating tumor cells. Radiol. Oncol. 2019, 53, 131–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Lorente, D.; Olmos, D.; Mateo, J.; Bianchini, D.; Seed, G.; Fleisher, M.; Danila, D.C.; Flohr, P.; Crespo, M.; Figueiredo, I.; et al. Decline in Circulating Tumor Cell Count and Treatment Outcome in Advanced Prostate Cancer. Eur. Urol. 2016, 70, 985–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grisanti, S.; Almici, C.; Consoli, F.; Buglione, M.; Verardi, R.; Bolzoni-Villaret, A.; Bianchetti, A.; Ciccarese, C.; Mangoni, M.; Ferrari, L.; et al. Circulating tumor cells in patients with recurrent or metastatic head and neck carcinoma: Prognostic and predictive significance. PLoS ONE 2014, 9, e103918. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [Green Version]
- Tayoun, T.; Faugeroux, V.; Oulhen, M.; Aberlenc, A.; Pawlikowska, P.; Farace, F. CTC-Derived Models: A Window into the Seeding Capacity of Circulating Tumor Cells (CTCs). Cells 2019, 8, 1145. [Google Scholar] [CrossRef] [Green Version]
- Holz, C.; Niehr, F.; Boyko, M.; Hristozova, T.; Distel, L.; Budach, V.; Tinhofer, I. Epithelial-mesenchymal-transition induced by EGFR activation interferes with cell migration and response to irradiation and cetuximab in head and neck cancer cells. Radiother. Oncol. 2011, 101, 158–164. [Google Scholar] [CrossRef]
- Durdik, M.; Kosik, P.; Gursky, J.; Vokalova, L.; Markova, E.; Belyaev, I. Imaging flow cytometry as a sensitive tool to detect low-dose-induced DNA damage by analyzing 53BP1 and γH2AX foci in human lymphocytes. Cytometry A 2015, 87, 1070–1078. [Google Scholar] [CrossRef]
- Keller, L.; Werner, S.; Pantel, K. Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165–180. [Google Scholar] [CrossRef] [Green Version]
- Rivera, F.; García-Castaño, A.; Vega, N.; Vega-Villegas, M.E.; Gutiérrez-Sanz, L. Cetuximab in metastatic or recurrent head and neck cancer: The EXTREME trial. Expert Rev. Anticancer Ther. 2009, 9, 1421–1428. [Google Scholar] [CrossRef]
- Ruiz-Rodríguez, A.J.; Molina-Vallejo, M.P.; Aznar-Peralta, I.; González Puga, C.; Cañas García, I.; González, E.; Lorente, J.A.; Serrano, M.J.; Garrido-Navas, M.C. Deep Phenotypic Characterisation of CTCs by Combination of Microfluidic Isolation (IsoFlux) and Imaging Flow Cytometry (ImageStream). Cancers 2021, 13, 6386. [Google Scholar] [CrossRef]
- Chudasama, D.; Katopodis, P.; Stone, N.; Haskell, J.; Sheridan, H.; Gardner, B.; Urnovitz, H.; Schuetz, E.; Beck, J.; Hall, M.; et al. Liquid Biopsies in Lung Cancer: Four Emerging Technologies and Potential Clinical Applications. Cancers 2019, 11, 331. [Google Scholar] [CrossRef] [Green Version]
- Dent, B.M.; Ogle, L.F.; O’Donnell, R.L.; Hayes, N.; Malik, U.; Curtin, N.J.; Boddy, A.V.; Plummer, E.R.; Edmondson, R.J.; Reeves, H.L.; et al. High-resolution imaging for the detection and characterisation of circulating tumour cells from patients with oesophageal, hepatocellular, thyroid and ovarian cancers. Int. J. Cancer 2016, 138, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Shirai, K.; Ijiri, Y.; Morita, E.; Yoshida, T.; Iwanaga, S.; Yanagida, M. Integrated system for detection and molecular characterization of circulating tumor cells. PLoS ONE 2020, 15, e0237506. [Google Scholar] [CrossRef]
- Ogle, L.F.; Orr, J.G.; Willoughby, C.E.; Hutton, C.; McPherson, S.; Plummer, R.; Boddy, A.V.; Curtin, N.J.; Jamieson, D.; Reeves, H.L. Imagestream detection and characterisation of circulating tumour cells—A liquid biopsy for hepatocellular carcinoma? J. Hepatol. 2016, 65, 305–313. [Google Scholar] [CrossRef] [Green Version]
- López-Riquelme, N.; Minguela, A.; Villar-Permuy, F.; Ciprian, D.; Castillejo, A.; Álvarez-López, M.R.; Soto, J.L. Imaging cytometry for counting circulating tumor cells: Comparative analysis of the CellSearch vs ImageStream systems. Apmis 2013, 121, 1139–1143. [Google Scholar] [CrossRef]
- Harris, E.J.; Huang, J.; Carroll, E.; Lowe, A.C.; Chau, N.G.; Rabinowits, G.; Haddad, R.; Hanna, G.J.; Haddad, T.; Sanborn, M.; et al. Circulating tumor cell analysis in locally advanced and metastatic squamous cell carcinoma of the head and neck. Laryngoscope Investig. Otolaryngol. 2020, 5, 1063–1069. [Google Scholar] [CrossRef]
- Rodemann, H.P.; Dittmann, K.; Toulany, M. Radiation-induced EGFR-signaling and control of DNA-damage repair. Int. J. Radiat. Biol. 2007, 83, 781–791. [Google Scholar] [CrossRef]
- Ang, K.K.; Berkey, B.A.; Tu, X.; Zhang, H.Z.; Katz, R.; Hammond, E.H.; Fu, K.K.; Milas, L. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002, 62, 7350–7356. [Google Scholar]
- Serrano, M.J.; Alvarez-Cubero, M.J.; De Miguel Pérez, D.; Rodríguez-Martínez, A.; Gonzalez-Herrera, L.; Robles-Fernandez, I.; Hernandez, J.E.; Puche, J.L.G.; Lorente, J.A. Significance of EGFR Expression in Circulating Tumor Cells. Adv. Exp. Med. Biol. 2017, 994, 285–296. [Google Scholar] [CrossRef]
- Tinhofer, I.; Hristozova, T.; Stromberger, C.; Keilhoiz, U.; Budach, V. Monitoring of circulating tumor cells and their expression of EGFR/phospho-EGFR during combined radiotherapy regimens in locally advanced squamous cell carcinoma of the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 2012, 83, e685–e690. [Google Scholar] [CrossRef]
- Carusillo, A.; Mussolino, C. DNA Damage: From Threat to Treatment. Cells 2020, 9, 1665. [Google Scholar] [CrossRef]
- Martin, O.A.; Anderson, R.L.; Russell, P.A.; Cox, R.A.; Ivashkevich, A.; Swierczak, A.; Doherty, J.P.; Jacobs, D.H.; Smith, J.; Siva, S.; et al. Mobilization of viable tumor cells into the circulation during radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2014, 88, 395–403. [Google Scholar] [CrossRef]
- Martin, O.A.; Anderson, R.L.; Narayan, K.; MacManus, M.P. Does the mobilization of circulating tumour cells during cancer therapy cause metastasis? Nat. Rev. Clin. Oncol. 2017, 14, 32–44. [Google Scholar] [CrossRef]
- Vilalta, M.; Rafat, M.; Graves, E.E. Effects of radiation on metastasis and tumor cell migration. Cell. Mol. Life Sci. 2016, 73, 2999–3007. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.L.; Adams, D.K.; He, J.; Kalhor, N.; Zhang, M.; Xu, T.; Gao, H.; Reuben, J.M.; Qiao, Y.; Komaki, R.; et al. Sequential Tracking of PD-L1 Expression and RAD50 Induction in Circulating Tumor and Stromal Cells of Lung Cancer Patients Undergoing Radiotherapy. Clin. Cancer Res. 2017, 23, 5948–5958. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, Z.G.; Yuan, H.; Deng, W.; Li, J.; Huang, Y.; Kim, B.Y.S.; Story, M.D.; Jiang, W. The Reciprocity between Radiotherapy and Cancer Immunotherapy. Clin. Cancer Res. 2019, 25, 1709–1717. [Google Scholar] [CrossRef] [Green Version]
- Gniadek, T.J.; Li, Q.K.; Tully, E.; Chatterjee, S.; Nimmagadda, S.; Gabrielson, E. Heterogeneous expression of PD-L1 in pulmonary squamous cell carcinoma and adenocarcinoma: Implications for assessment by small biopsy. Mod. Pathol. 2017, 30, 530–538. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, J.H.; Lelkaitis, G.; Håkansson, K.; Vogelius, I.R.; Johannesen, H.H.; Fischer, B.M.; Bentzen, S.M.; Specht, L.; Kristensen, C.A.; von Buchwald, C.; et al. Intratumor heterogeneity of PD-L1 expression in head and neck squamous cell carcinoma. Br. J. Cancer 2019, 120, 1003–1006. [Google Scholar] [CrossRef] [Green Version]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khattak, M.A.; Reid, A.; Freeman, J.; Pereira, M.; McEvoy, A.; Lo, J.; Frank, M.H.; Meniawy, T.; Didan, A.; Spencer, I.; et al. PD-L1 Expression on Circulating Tumor Cells May Be Predictive of Response to Pembrolizumab in Advanced Melanoma: Results from a Pilot Study. Oncologist 2020, 25, e520–e527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janning, M.; Kobus, F.; Babayan, A.; Wikman, H.; Velthaus, J.-L.; Bergmann, S.; Schatz, S.; Falk, M.; Berger, L.-A.; Böttcher, L.-M.; et al. Determination of PD-L1 Expression in Circulating Tumor Cells of NSCLC Patients and Correlation with Response to PD-1/PD-L1 Inhibitors. Cancers 2019, 11, 835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibaki, R.; Koh, Y.; Akamatsu, H.; Kurita, K.; Yagi, S.; Kanai, K.; Hayata, A.; Tokudome, N.; Higuchi, M.; Kanbara, H.; et al. Predictive impact of PD-L1-expressing circulating tumor cells in NSCLC patients treated with nivolumab. J. Clin. Oncol. 2017, 35, 11541. [Google Scholar] [CrossRef]
- Nicolazzo, C.; Raimondi, C.; Mancini, M.; Caponnetto, S.; Gradilone, A.; Gandini, O.; Mastromartino, M.; Del Bene, G.; Prete, A.; Longo, F.; et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci. Rep. 2016, 6, 31726. [Google Scholar] [CrossRef] [PubMed]
- Strati, A.; Koutsodontis, G.; Papaxoinis, G.; Angelidis, I.; Zavridou, M.; Economopoulou, P.; Kotsantis, I.; Avgeris, M.; Mazel, M.; Perisanidis, C.; et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 2017, 28, 1923–1933. [Google Scholar] [CrossRef]
- Tan, Z.; Yue, C.; Ji, S.; Zhao, C.; Jia, R.; Zhang, Y.; Liu, R.; Li, D.; Yu, Q.; Li, P.; et al. Assessment of PD-L1 Expression on Circulating Tumor Cells for Predicting Clinical Outcomes in Patients with Cancer Receiving PD-1/PD-L1 Blockade Therapies. Oncologist 2021, 26, e2227–e2238. [Google Scholar] [CrossRef]
- Man, J.; Millican, J.; Mulvey, A.; Gebski, V.; Hui, R. Response Rate and Survival at Key Timepoints With PD-1 Blockade vs Chemotherapy in PD-L1 Subgroups: Meta-Analysis of Metastatic NSCLC Trials. JNCI Cancer Spectr. 2021, 5, pkab012. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Perry, C.; Warkiani, M.E.; Blick, T.; Davies, A.; O’Byrne, K.; Thompson, E.W.; Nelson, C.C.; Vela, I.; Punyadeera, C. Short term ex-vivo expansion of circulating head and neck tumour cells. Oncotarget 2016, 7, 60101–60109. [Google Scholar] [CrossRef] [Green Version]
- Koh, Y.; Yagi, S.; Akamatsu, H.; Kanai, K.; Hayata, A.; Tokudome, N.; Akamatsu, K.; Higuchi, M.; Kanbara, H.; Nakanishi, M.; et al. Heterogeneous Expression of Programmed Death Receptor-ligand 1 on Circulating Tumor Cells in Patients With Lung Cancer. Clin. Lung Cancer 2019, 20, 270–277.e271. [Google Scholar] [CrossRef] [Green Version]
- Ilié, M.; Szafer-Glusman, E.; Hofman, V.; Chamorey, E.; Lalvée, S.; Selva, E.; Leroy, S.; Marquette, C.H.; Kowanetz, M.; Hedge, P.; et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann. Oncol. 2018, 29, 193–199. [Google Scholar] [CrossRef]
- Evrard, D.; Hourseau, M.; Couvelard, A.; Paradis, V.; Gauthier, H.; Raymond, E.; Halimi, C.; Barry, B.; Faivre, S. PD-L1 expression in the microenvironment and the response to checkpoint inhibitors in head and neck squamous cell carcinoma. Oncoimmunology 2020, 9, 1844403. [Google Scholar] [CrossRef]
- Tu, X.; Qin, B.; Zhang, Y.; Zhang, C.; Kahila, M.; Nowsheen, S.; Yin, P.; Yuan, J.; Pei, H.; Li, H.; et al. PD-L1 (B7-H1) Competes with the RNA Exosome to Regulate the DNA Damage Response and Can Be Targeted to Sensitize to Radiation or Chemotherapy. Mol. Cell 2019, 74, 1215–1226.e1214. [Google Scholar] [CrossRef]
- Satelli, A.; Batth, I.S.; Brownlee, Z.; Rojas, C.; Meng, Q.H.; Kopetz, S.; Li, S. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci. Rep. 2016, 6, 28910. [Google Scholar] [CrossRef]





| HNSCC | BC | ||
|---|---|---|---|
| gender | |||
| n | female | 4 | 8 |
| male | 12 | - | |
| age (years) | |||
| median (range) | female | 70 (32–81) | 48 (34–64) |
| male | 69 (58–79) | - | |
| stage of disease | |||
| n (%) | early stage | - | 4 (50%) |
| locally advanced | 1 (6%) | - | |
| recurrent/metastatic | 15 (94%) | 4 (50%) | |
| tumor site | |||
| n (%) | oral cavity | 8 (50%) | - |
| oropharynx | 3 (19%) | - | |
| hypopharynx | 3 (19%) | - | |
| other/breast | 2 (12%) | 8 (100%) | |
| metastatic sites | |||
| n (%) | none | 1 (0.06%) | 4 (50%) |
| regional | 3 (25%) | 3 (38%) | |
| distant | 12 (75%) | 1(12%) | |
| CTCpositive cases (≥ 3 CTCs) | |||
| n (%) | 7 (44%) | 6 (75%) | |
| CTC numbers | |||
| median | 15 | 14 | |
| range | 6–30 | 9–27 | |
| PD-L1positive cases | |||
| n (%) | 4 (57%) | 4 (67%) | |
| PD-L1positive cells | |||
| median (n) | 6 | 2 | |
| range (n) | 3–30 | 1–6 | |
| % of PD-L1positive CTCs in entire CTC population | median (%) range (%) | 100% 56%–100% | 15% 4%–25% |
| Pat.ID | Tumor Site | Date of tissue Biopsy (Month/Year) | Date of Liquid Biopsy (Month/Year) | PD–L1positive Cells in Tumor Tissue (TPS %) | PD–L1positive CTCs (%) | Time between Tumor and Liquid Biopsy (Months) |
|---|---|---|---|---|---|---|
| BC-003 | breast | December/2017 | December /2017 | 0 | 8 | 0.4 |
| BC-006 | breast | December /2017 | January/2018 | 0 | 20 | 1 |
| HNC-012 | oral cavity | September/2017 | September /2017 | 70 | 100 | 0.4 |
| HNC-018 | oral cavity | July/2016 | January /2018 | 0 | 0 | 19 |
| HNC-019 | oral cavity | September /2017 | January /2018 | 0 | 45 | 5 |
| HNC-020 | hypopharynx | January/2017 | January /2018 | 5 | 100 | 12 |
| HNC-026 | hypopharynx | December./2016 | Mar./2018 | 0 | 0 | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staudte, S.; Klinghammer, K.; Jurmeister, P.S.; Jank, P.; Blohmer, J.-U.; Liebs, S.; Rhein, P.; Hauser, A.E.; Tinhofer, I. Multiparametric Phenotyping of Circulating Tumor Cells for Analysis of Therapeutic Targets, Oncogenic Signaling Pathways and DNA Repair Markers. Cancers 2022, 14, 2810. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14112810
Staudte S, Klinghammer K, Jurmeister PS, Jank P, Blohmer J-U, Liebs S, Rhein P, Hauser AE, Tinhofer I. Multiparametric Phenotyping of Circulating Tumor Cells for Analysis of Therapeutic Targets, Oncogenic Signaling Pathways and DNA Repair Markers. Cancers. 2022; 14(11):2810. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14112810
Chicago/Turabian StyleStaudte, Stephanie, Konrad Klinghammer, Philipp Sebastian Jurmeister, Paul Jank, Jens-Uwe Blohmer, Sandra Liebs, Peter Rhein, Anja E. Hauser, and Ingeborg Tinhofer. 2022. "Multiparametric Phenotyping of Circulating Tumor Cells for Analysis of Therapeutic Targets, Oncogenic Signaling Pathways and DNA Repair Markers" Cancers 14, no. 11: 2810. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14112810