Routine EWS Fusion Analysis in the Oncology Clinic to Identify Cancer-Specific Peptide Sequence Patterns That Span Breakpoints in Ewing Sarcoma and DSRCT
Abstract
:Simple Summary
Abstract
1. Introduction
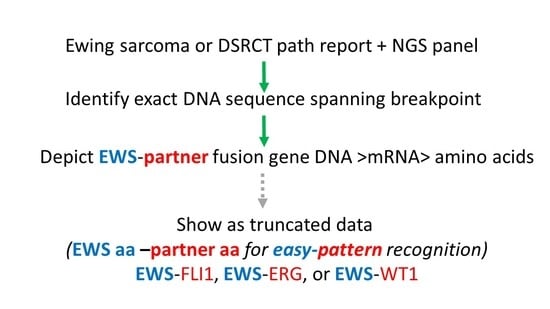
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Womer, R.B.; West, D.C.; Krailo, M.D.; Dickman, P.S.; Pawel, B.R.; Grier, H.E.; Marcus, K.; Sailer, S.; Healey, J.H.; Dormans, J.P.; et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: A report from the Children’s Oncology Group. J. Clin. Oncol. 2012, 30, 4148–4154. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.-C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riggi, N.; Suva, M.; Stamenkovic, I. Ewing’s Sarcoma. N. Engl. J. Med. 2021, 384, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Granowetter, L.; Womer, R.; Devidas, M.; Krailo, M.; Wang, C.; Bernstein, M.; Marina, N.; Leavey, P.; Gebhardt, M.; Healey, J.; et al. Dose-intensified compared with standard chemotherapy for nonmetastatic Ewing sarcoma family of tumors: A Children’s Oncology Group Study. J. Clin. Oncol. 2009, 27, 2536–2541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dirksen, U.; Brennan, B.; Le Deley, M.-C.; Cozic, N.; Van Den Berg, H.; Bhadri, V.; Brichard, B.; Claude, L.; Craft, A.; Amler, S.; et al. High-Dose Chemotherapy Compared With Standard Chemotherapy and Lung Radiation in Ewing Sarcoma With Pulmonary Metastases: Results of the European Ewing Tumour Working Initiative of National Groups, 99 Trial and EWING 2008. J. Clin. Oncol. 2019, 37, 3192–3202. [Google Scholar] [CrossRef]
- Whelan, J.; Le Deley, M.-C.; Dirksen, U.; Le Teuff, G.; Brennan, B.; Gaspar, N.; Hawkins, D.S.; Amler, S.; Bauer, S.; Bielack, S.; et al. High-Dose Chemotherapy and Blood Autologous Stem-Cell Rescue Compared With Standard Chemotherapy in Localized High-Risk Ewing Sarcoma: Results of Euro-E.W.I.N.G.99 and Ewing-2008. J. Clin. Oncol. 2018, 36, 3110–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasper, M.; Jabar, S.; Ranft, A.; Jürgens, H.; Amler, S.; Dirksen, U. The value of high-dose chemotherapy in patients with first relapsed Ewing sarcoma. Pediatr. Blood Cancer 2014, 61, 1382–1386. [Google Scholar] [CrossRef] [PubMed]
- Windsor, R.; Hamilton, A.; McTiernan, A.; Dileo, P.; Michelagnoli, M.; Seddon, B.; Strauss, S.J.; Whelan, J. Survival after high-dose chemotherapy for refractory and recurrent Ewing sarcoma. Eur. J. Cancer 2022, 170, 131–139. [Google Scholar] [CrossRef]
- Mackall, C.L.; Rhee, E.H.; Read, E.J.; Khuu, H.M.; Leitman, S.F.; Bernstein, D.; Tesso, M.; Long, L.M.; Grindler, D.; Merino, M.; et al. A pilot study of consolidative immunotherapy in patients with high-risk pediatric sarcomas. Clin. Cancer Res. 2008, 14, 4850–4858. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.K.; Witten, B.G.; Harmsen, W.S.; Rose, P.S.; Krailo, M.; Marcus, K.J.; Randall, R.L.; DuBois, S.G.; Janeway, K.A.; Womer, R.B.; et al. Ewing’s sarcoma: Only patients with 100% of necrosis after chemotherapy should be classified as having a good response. Bone Joint J. 2016, 98, 1138–1144. [Google Scholar]
- Bosma, S.E.; Lancia, C.; Rueten-Budde, A.J.; Ranft, A.; Gelderblom, H.; Fiocco, M.; van de Sande, M.A.J.; Dijkstra, P.D.S.; Dirksen, U. Easy-to-use clinical tool for survival estimation in Ewing sarcoma at diagnosis and after surgery. Sci. Rep. 2019, 9, 11000. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.K.; Witten, B.G.; Harmsen, W.S.; Rose, P.S.; Krailo, M.; Marcus, K.J.; Randall, R.L.; DuBois, S.G.; A Janeway, K.; Womer, R.B.; et al. Analysis of local control outcomes and clinical prognostic factors in localized pelvic Ewing sarcoma patients treated with radiation therapy: A Report from the Children’s Oncology Group. Int. J. Radiat. Oncol. Biol. Phys. 2022, 115, 337–346. [Google Scholar] [CrossRef]
- Lozano-Calderón, S.A.; Albergo, J.I.; Groot, O.Q.; Merchan, N.A.; El Abiad, J.M.; Salinas, V.; Mier, L.C.G.; Montoya, C.S.; Ferrone, M.L.; Ready, J.E.; et al. Complete tumor necrosis after neoadjuvant chemotherapy defines good responders in patients with Ewing sarcoma. Cancer 2022, 129, 60–70. [Google Scholar] [CrossRef]
- Stahl, M.; Ranft, A.; Paulussen, M.; Bölling, T.; Vieth, V.; Bielack, S.; Görtitz, I.; Braun-Munzinger, G.; Hardes, J.; Jürgens, H.; et al. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr. Blood Cancer 2011, 57, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Bosma, S.E.; Ayu, O.; Fiocco, M.; Gelderblom, H.; Dijkstra, P.D.S. Prognostic factors for survival in Ewing sarcoma: A systematic review. Surg. Oncol. 2018, 27, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Alava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Primers. 2018, 4, 5. [Google Scholar] [CrossRef]
- A Showpnil, I.; Selich-Anderson, J.; Taslim, C.; A Boone, M.; Crow, J.C.; Theisen, E.R.; Lessnick, S.L. EWS/FLI mediated reprogramming of 3D chromatin promotes an altered transcriptional state in Ewing sarcoma. Nucleic Acids Res. 2022, 50, 9814–9837. [Google Scholar] [CrossRef] [PubMed]
- Shulman, D.S.; Whittle, S.B.; Surdez, D.; Bailey, K.M.; de Álava, E.; Yustein, J.T.; Shlien, A.; Hayashi, M.; Bishop, A.J.R.; Crompton, B.D.; et al. An international working group consensus report for the prioritization of molecular biomarkers for Ewing sarcoma. NPJ Precis. Oncol. 2022, 6, 65. [Google Scholar] [CrossRef]
- Apfelbaum, A.A.; Wu, F.; Hawkins, A.G.; Magnuson, B.; Jiménez, J.A.; Taylor, S.D.; Wrenn, E.D.; Waltner, O.; Pfaltzgraff, E.R.; Song, J.Y.; et al. EWS: FLI1 and HOXD13 Control Tumor Cell Plasticity in Ewing Sarcoma. Clin. Cancer Res. 2022, 28, 4466–4478. [Google Scholar] [CrossRef]
- Weaver, D.T.; Pishas, K.I.; Williamson, D.; Scarborough, J.; Lessnick, S.L.; Dhawan, A.; Scott, J.G. Network potential identifies therapeutic miRNA cocktails in Ewing sarcoma. PLoS Comput. Biol. 2021, 17, e1008755. [Google Scholar] [CrossRef]
- Boone, M.A.; Taslim, C.; Crow, J.C.; Selich-Anderson, J.; Byrum, A.K.; Showpnil, I.A.; Sunkel, B.D.; Wang, M.; Stanton, B.Z.; Theisen, E.R.; et al. The FLI portion of EWS/FLI contributes a transcriptional regulatory function that is distinct and separable from its DNA-binding function in Ewing sarcoma. Oncogene 2021, 40, 4759–4769. [Google Scholar] [CrossRef]
- Deng, Q.; Natesan, R.; Cidre-Aranaz, F.; Arif, S.; Liu, Y.; Rasool, R.U.; Wang, P.; Mitchell-Velasquez, E.; Das, C.K.; Vinca, E.; et al. Oncofusion-driven de novo enhancer assembly promotes malignancy in Ewing sarcoma via aberrant expression of the stereociliary protein LOXHD1. Cell Rep. 2022, 39, 110971. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.A.; Apfelbaum, A.A.; Hawkins, A.G.; Svoboda, L.K.; Kumar, A.; Ruiz, R.O.; Garcia, A.X.; Haarer, E.; Nwosu, Z.C.; Bradin, J.; et al. EWS-FLI1 and Menin Converge to Regulate ATF4 Activity in Ewing Sarcoma. Mol. Cancer Res. 2021, 19, 1182–1195. [Google Scholar] [CrossRef]
- Theisen, E.R.; Miller, K.R.; Showpnil, I.A.; Taslim, C.; Pishas, K.I.; Lessnick, S.L. Transcriptomic analysis functionally maps the intrinsically disordered domain of EWS/FLI and reveals novel transcriptional dependencies for oncogenesis. Genes Cancer 2019, 10, 21–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, A.G.; Basrur, V.; Leprevost, F.D.V.; Pedersen, E.; Sperring, C.; Nesvizhskii, A.I.; Lawlor, E.R. The Ewing Sarcoma Secretome and Its Response to Activation of Wnt/beta-catenin Signaling. Mol. Cell. Proteom. 2018, 17, 901–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovar, H.; Amatruda, J.; Brunet, E.; Burdach, S.; Cidre-Aranaz, F.; de Alava, E.; Dirksen, U.; van der Ent, W.; Grohar, P.; Grünewald, T.G.P.; et al. The second European interdisciplinary Ewing sarcoma research summit--A joint effort to deconstructing the multiple layers of a complex disease. Oncotarget 2016, 7, 8613–8624. [Google Scholar] [CrossRef] [Green Version]
- Scopim-Ribeiro, R.; Lizardo, M.M.; Zhang, H.-F.; Dhez, A.-C.; Hughes, C.S.; Sorensen, P.H. NSG Mice Facilitate ex vivo Characterization of Ewing Sarcoma Lung Metastasis Using the PuMA Model. Front.Oncol. 2021, 11, 645757. [Google Scholar] [CrossRef]
- Liu, H.; Huang, L.; Luo, J.; Chen, W.; Zhang, Z.; Liao, X.; Dai, M.; Shu, Y.; Cao, K. Prediction and identification of B cell epitopes derived from EWS/FLI-l fusion protein of Ewing’s sarcoma. Med. Oncol. 2012, 29, 3421–3430. [Google Scholar] [CrossRef]
- Mackall, C.L. In search of targeted therapies for childhood cancer. Front. Oncol. 2011, 1, 18. [Google Scholar] [CrossRef] [Green Version]
- Orentas, R.J.; Lee, D.; Mackall, C. Immunotherapy targets in pediatric cancer. Front. Oncol. 2012, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Kilpatrick, S.E.; Reith, J.D.; Rubin, B. Ewing Sarcoma and the History of Similar and Possibly Related Small Round Cell Tumors: From Whence Have We Come and Where are We Going? Adv. Anat. Pathol. 2018, 25, 314–326. [Google Scholar] [CrossRef]
- Cheng, Y.-W.; Meyer, A.; A Jakubowski, M.; O Keenan, S.; E Brock, J.; Azzato, E.M.; Weindel, M.; Farkas, D.H.; Rubin, B.P. Gene Fusion Identification Using Anchor-Based Multiplex PCR and Next-Generation Sequencing. J. Appl. Lab. Med. 2021, 6, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.M.; Trucco, M.M.; Garzone, S.; Sartoski, S.; Nystrom, L.; Zahler, S.; Thomas, S.; Murphy, E.S. Virtual visits for sarcomas and other rare cancers including desmoids: The evolution of informaton and educational opportunities to improve health: 2017–2022. In Proceedings of the Connective Tissue Oncology Society (CTOS), Vancouver, BC, Canada, 16–19 November 2022. [Google Scholar]
- Anderson, P.M.; Hanna, R. Defining Moments: Making Time for Virtual Visits and Catalyzing Better Cancer Care. Health Commun. 2020, 35, 787–791. [Google Scholar] [CrossRef]
- Le Deley, M.-C.; Delattre, O.; Schaefer, K.-L.; Burchill, S.A.; Koehler, G.; Hogendoorn, P.C.; Lion, T.; Poremba, C.; Marandet, J.; Ballet, S.; et al. Impact of EWS-ETS fusion type on disease progression in Ewing’s sarcoma/peripheral primitive neuroectodermal tumor: Prospective results from the cooperative Euro-E.W.I.N.G. 99 trial. J. Clin. Oncol. 2010, 28, 1982–1988. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lee, K.-W.; Srivastava, R.M.; Kuo, F.; Krishna, C.; Chowell, D.; Makarov, V.; Hoen, D.; Dalin, M.G.; Wexler, L.; et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat. Med. 2019, 25, 767–775. [Google Scholar] [CrossRef]
- Palmer, C.D.; Rappaport, A.R.; Davis, M.J.; Hart, M.G.; Scallan, C.D.; Hong, S.-J.; Gitlin, L.; Kraemer, L.D.; Kounlavouth, S.; Yang, A.; et al. Individualized, heterologous chimpanzee adenovirus and self-amplifying mRNA neoantigen vaccine for advanced metastatic solid tumors: Phase 1 trial interim results. Nat. Med. 2022, 28, 1619–1629. [Google Scholar] [CrossRef]
- Sellars, M.C.; Wu, C.J.; Fritsch, E.F. Cancer vaccines: Building a bridge over troubled waters. Cell 2022, 185, 2770–2788. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Graham, G.T.; Selvanathan, S.P.; Zöllner, S.K.; Stahl, E.; Shlien, A.; Caplen, N.J.; Üren, A.; A Toretsky, J. Comprehensive profiling of mRNA splicing indicates that GC content signals altered cassette exon inclusion in Ewing sarcoma. NAR Cancer 2022, 4, zcab052. [Google Scholar] [CrossRef]
- Flores, G.; Grohar, P.J. One oncogene, several vulnerabilities: EWS/FLI targeted therapies for Ewing sarcoma. J. Bone Oncol. 2021, 31, 100404. [Google Scholar] [CrossRef] [PubMed]
- Pishas, K.I.; Drenberg, C.D.; Taslim, C.; Theisen, E.R.; Johnson, K.M.; Saund, R.S.; Pop, I.L.; Crompton, B.D.; Lawlor, E.R.; Tirode, F.; et al. Therapeutic Targeting of KDM1A/LSD1 in Ewing Sarcoma with SP-2509 Engages the Endoplasmic Reticulum Stress Response. Mol. Cancer Ther. 2018, 17, 1902–1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harlow, M.L.; Maloney, N.; Roland, J.; Navarro, M.J.G.; Easton, M.K.; Kitchen-Goosen, S.M.; Boguslawski, E.A.; Madaj, Z.B.; Johnson, B.K.; Bowman, M.J.; et al. Lurbinectedin Inactivates the Ewing Sarcoma Oncoprotein EWS-FLI1 by Redistributing It within the Nucleus. Cancer Res. 2016, 76, 6657–6668. [Google Scholar] [CrossRef] [Green Version]
- Volchenboum, S.L.; Andrade, J.; Huang, L.; Barkauskas, D.A.; Krailo, M.; Womer, R.B.; Ranft, A.; Potratz, J.; Dirksen, U.; Triche, T.J.; et al. Gene Expression Profiling of Ewing Sarcoma Tumors Reveals the Prognostic Importance of Tumor-Stromal Interactions: A Report from the Children’s Oncology Group. J. Pathol. Clin. Res. 2015, 1, 83–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, M.M.Y.; Maleki Vareki, S. Addressing the Elephant in the Immunotherapy Room: Effector T-Cell Priming versus Depletion of Regulatory T-Cells by Anti-CTLA-4 Therapy. Cancers 2022, 14, 1580. [Google Scholar] [CrossRef]
- Li, Y.-J.; Yang, X.; Zhang, W.-B.; Yi, C.; Wang, F.; Li, P. Clinical implications of six inflammatory biomarkers as prognostic indicators in Ewing sarcoma. Cancer Manag. Res. 2017, 9, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Gassmann, H.; Schneider, K.; Evdokimova, V.; Ruzanov, P.; Schober, S.J.; Xue, B.; von Heyking, K.; Thiede, M.; Richter, G.H.S.; Pfaffl, M.W.; et al. Ewing Sarcoma-Derived Extracellular Vesicles Impair Dendritic Cell Maturation and Function. Cells 2021, 10, 2081. [Google Scholar] [CrossRef]
- Subbiah, V. The next generation of evidence-based medicine. Nat. Med. 2023, 29, 49–58. [Google Scholar] [CrossRef]




| EWS-FLI1 Breakpoint Fusion Peptide Analysis | Number (Per Cent) |
|---|---|
| SQQSSSYGQQNPSYDSVRRG | 19 |
| SQQSSSYGQQSSLLAYNTTS | 12 |
| SQQSSSYGQQNPYQILGPTS | 1 |
| SQQSSSYGQQRSGQIQLWQF | 1 |
| 33/40 (83%) | |
| Other EWS-FLI1 Fusions Peptides | |
| PMDEGPDLDLGSLLAYNTTS | 5 |
| GERGGFNKPGGPPLGGAQTI | 1 |
| CVEFSSLIDQPVYPDVLASG | 1 |
| 7/40 (17%) |
| Ewing Sarcomas with EWS-ERG Fusions |
|---|
| EWS-ERG exon 7-9 (N = 3/6, 50%) SQQSSSYGQQNLPYEPPRRS |
| Ewing sarcomas with EWS-FEV fusions |
| EWS-FEV1 exon 7-2 (2/3; 67%) SQQSSSYGQQNPVGDGLFKD |
| DSRCT with EWS-WT1 fusions |
| EWS-WT1 exon 7-7 (N = 9/12, 75%) SQQSSSYGQQSEKPYQCDFK |
| EWS-WT1 exon 9-7 (N = 3/12 25%) GERGGFNKPGGEKPYQCDFK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, P.M.; Tu, Z.J.; Kilpatrick, S.E.; Trucco, M.; Hanna, R.; Chan, T. Routine EWS Fusion Analysis in the Oncology Clinic to Identify Cancer-Specific Peptide Sequence Patterns That Span Breakpoints in Ewing Sarcoma and DSRCT. Cancers 2023, 15, 1623. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers15051623
Anderson PM, Tu ZJ, Kilpatrick SE, Trucco M, Hanna R, Chan T. Routine EWS Fusion Analysis in the Oncology Clinic to Identify Cancer-Specific Peptide Sequence Patterns That Span Breakpoints in Ewing Sarcoma and DSRCT. Cancers. 2023; 15(5):1623. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers15051623
Chicago/Turabian StyleAnderson, Peter M., Zheng Jin Tu, Scott E. Kilpatrick, Matteo Trucco, Rabi Hanna, and Timothy Chan. 2023. "Routine EWS Fusion Analysis in the Oncology Clinic to Identify Cancer-Specific Peptide Sequence Patterns That Span Breakpoints in Ewing Sarcoma and DSRCT" Cancers 15, no. 5: 1623. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers15051623