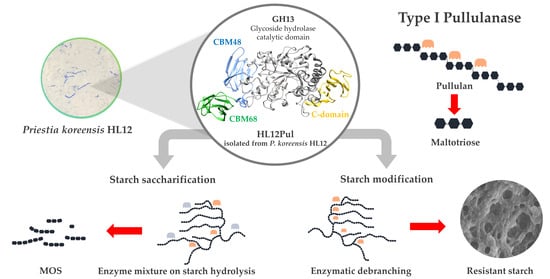
In-Depth Characterization of Debranching Type I Pullulanase from Priestia koreensis HL12 as Potential Biocatalyst for Starch Saccharification and Modification
Abstract
:1. Introduction
2. Results
2.1. Pullulanase Gene Identification from P. koreensis HL12 and Sequence Analysis
2.2. Heterologous Expression and Purification of Pullulanase
2.3. Biochemical Characterization
2.4. Cleavage Pattern on Pullulan and Specific Linkages
2.5. Potential of HL12Pul in Raw Starch Saccharification
2.6. Enzymatic Process for Resistant Starch Production
2.7. In Vitro Digestibility of Modified Starch
3. Discussions
4. Materials and Methods
4.1. Chemicals, Bacterial Strains and Plasmids
4.2. Identification of HL12PUL Type I Pullulanase Gene from P. koeensis HL12
4.3. Heterologous Expression and Purification of Pullulanase
4.4. Purification of Recombinant HL12Pul
4.5. Enzyme Activity Assay
4.6. Determination of Biochemical Properties of Recombinant HL12Pul
4.7. Determination of Kinetic Parameters on Pullulan
4.8. Verification of HL12Pul Specific Cleavage of Glycosidic Linkages
4.9. Synergistic Effects of HL12Pul in Starch Saccharification
4.10. Enzymatic Process for Resistant Starch Production
4.11. Physiochemical and Biochemical Properties Evaluation of Modified Starch
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hii, S.L.; Tan, J.S.; Ling, T.C.; Ariff, A.B. Pullulanase: Role in starch hydrolysis and potential industrial applications. Enzym. Res. 2012, 2012, 921362. [Google Scholar] [CrossRef]
- Alcázar-Alay, S.C.; Meireles, M.A. Physicochemical properties, modifications and applications of starches from different botanical sources. Food Sci. Technol. 2015, 35, 215–236. [Google Scholar] [CrossRef]
- Xu, J.; Ren, F.; Huang, C.-H.; Zheng, Y.; Zhen, J.; Sun, H.; Ko, T.-P.; He, M.; Chen, C.-C.; Chan, H.-C.; et al. Functional and structural studies of pullulanase from Anoxybacillus sp. LM18-11. Proteins 2014, 82, 1685–1693. [Google Scholar] [CrossRef]
- Norman, B.E. A novel debranching enzyme for application in the glucose syrup industry. Starch-Stärke 1982, 34, 340–346. [Google Scholar] [CrossRef]
- Roy, I.; Gupta, M.N. Hydrolysis of starch by a mixture of glucoamylase and pullulanase entrapped individually in calcium alginate beads. Enzym. Microb. Technol. 2004, 34, 26–32. [Google Scholar] [CrossRef]
- Long, J.; Zhang, B.; Li, X.; Zhan, X.; Xu, X.; Xie, Z.; Jin, Z. Effective production of resistant starch using pullulanase immobilized onto magnetic chitosan/Fe3O4 nanoparticles. Food Chem. 2018, 239, 276–286. [Google Scholar] [CrossRef]
- Pongjanta, J.; Uthaipattanaceep, A.; Naivikul, O.; Piyachomkwan, K. Enzymes-resistant starch (RS III) from pullulanase- debranched high amylose rice starch. Kasetsart J. 2008, 42, 198–205. [Google Scholar]
- Kahar, U.M.; Latif, N.A.; Amran, S.I.; Liew, K.J.; Goh, K.M. A bibliometric analysis and review of pullulan-degrading enzymes—past and current trends. Catalysts 2022, 12, 143. [Google Scholar] [CrossRef]
- Møller, M.S.; Henriksen, A.; Svensson, B. Structure and function of α-glucan debranching enzymes. Cell. Mol. Life Sci. 2016, 73, 2619–2641. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Sharma, N.; Rai, A.K.; Singh, S.P. A novel cold-active type I pullulanase from a hot-spring metagenome for effective debranching and production of resistant starch. Bioresour. Technol. 2021, 320, 124288. [Google Scholar] [CrossRef] [PubMed]
- Janeček, Š.; Mareček, F.; MacGregor, E.A.; Svensson, B. Starch-binding domains as CBM families–history, occurrence, structure, function and evolution. Biotechnol. Adv. 2019, 37, 107451. [Google Scholar] [CrossRef] [PubMed]
- Iqrar, U.; Javaid, H.; Ashraf, N.; Ahmad, A.; Latief, N.; Shahid, A.A.; Ahmad, W.; Ijaz, B. Structural and functional analysis of pullulanase type 1 (PulA) from Geobacillus thermopakistaniensis. Mol. Biotechnol. 2020, 62, 370–379. [Google Scholar] [CrossRef]
- Xia, W.; Zhang, K.; Su, L.; Wu, J. Microbial starch debranching enzymes: Developments and applications. Biotechnol. Adv. 2021, 50, 107786. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, S.-Y.; Luo, Z.-G.; Zong, M.-H.; Li, X.-X.; Lou, W.-Y. Biotechnology and bioengineering of pullulanase: State of the art and perspectives. World J. Microbiol. Biotechnol. 2021, 37, 43. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: Description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar] [CrossRef]
- Lim, J.-M.; Jeon, C.O.; Lee, J.-C.; Ju, Y.J.; Park, D.-J.; Kim, C.-J. Bacillus koreensis sp. nov., a spore-forming bacterium, isolated from the rhizosphere of willow roots in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 59–63. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef]
- Janeček, Š.; Gabriško, M. Remarkable evolutionary relatedness among the enzymes and proteins from the α-amylase family. Cell. Mol. Life Sci. 2016, 73, 2707–2725. [Google Scholar] [CrossRef] [PubMed]
- Saburi, W.; Rachi-Otsuka, H.; Hondoh, H.; Okuyama, M.; Mori, H.; Kimura, A. Structural elements responsible for the glucosidic linkage-selectivity of a glycoside hydrolase family 13 exo-glucosidase. FEBS Lett. 2015, 589, 865–869. [Google Scholar] [CrossRef]
- Ito, K.; Ito, S.; Ishino, K.; Shimizu-Ibuka, A.; Sakai, H. Val326 of Thermoactinomyces vulgaris R-47 amylase II modulates the preference for alpha-(1,4)- and alpha-(1,6)-glycosidic linkages. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2007, 1774, 443–449. [Google Scholar] [CrossRef]
- Turkenburg, J.P.; Brzozowski, A.M.; Svendsen, A.; Borchert, T.V.; Davies, G.J.; Wilson, K.S. Structure of a pullulanase from Bacillus acidopullulyticus. Proteins Struct. Funct. Bioinform. 2009, 76, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Beatty, M.K.; Rahman, A.; Cao, H.; Woodman, W.; Lee, M.; Myers, A.M.; James, M.G. Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize1. Plant Physiol. 1999, 119, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Domań-Pytka, M.; Bardowski, J. Pullulan degrading enzymes of bacterial origin. Crit. Rev. Microbiol. 2004, 30, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ma, J.; Chen, S.-Q.; Cai, X.-H.; Wei, D.-Z. A novel cold-adapted type I pullulanase of Paenibacillus polymyxa Nws-pp2: In vivo functional expression and biochemical characterization of glucans hydrolyzates analysis. BMC Biotechnol. 2015, 15, 96. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzym. Microb. Technol. 2011, 49, 429–440. [Google Scholar] [CrossRef]
- Machovič, M.; Janeček, Š. Domain evolution in the GH13 pullulanase subfamily with focus on the carbohydrate-binding module family 48. Biologia 2008, 63, 1057–1068. [Google Scholar] [CrossRef]
- Zeng, Y.; Zheng, H.; Shen, Y.; Xu, J.; Tan, M.; Liu, F.; Song, H. Identification and analysis of binding residues in the CBM68 of pullulanase PulA from Anoxybacillus sp. LM18-11. J. Biosci. Bioeng. 2019, 127, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Janeček, Š.; Majzlová, K.; Svensson, B.; MacGregor, E.A. The starch-binding domain family CBM41—An in silico analysis of evolutionary relationships. Proteins Struct. Funct. Genet. 2017, 85, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Xu, J.; Fu, X.; Tan, M.; Liu, F.; Zheng, H.; Song, H. Effects of different carbohydrate-binding modules on the enzymatic properties of pullulanase. Int. J. Biol. Macromol. 2019, 137, 973–981. [Google Scholar] [CrossRef]
- Kahar, U.M.; Ng, C.L.; Chan, K.-G.; Goh, K.M. Characterization of a type I pullulanase from Anoxybacillus sp. SK3-4 reveals an unusual substrate hydrolysis. Appl. Microbiol. Biotechnol. 2016, 100, 6291–6307. [Google Scholar] [CrossRef]
- Rajaei, S.; Noghabi, K.A.; Sadeghizadeh, M.; Zahiri, H.S. Characterization of a pH and detergent-tolerant, cold-adapted type I pullulanase from Exiguobacterium sp. SH3. Extremophiles 2015, 19, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yan, Q.; Bao, Q.; Liu, J.; Jiang, Z. Expression and biochemical characterization of a novel type I pullulanase from Bacillus megaterium. Biotechnol. Lett. 2017, 39, 397–405. [Google Scholar] [CrossRef]
- Pang, B.; Zhou, L.; Cui, W.; Liu, Z.; Zhou, Z. Improvement of the thermostability and activity of pullulanase from Anoxybacillus sp. WB42. Appl. Biochem. Biotechnol. 2020, 191, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, Y.; Obaroakpo, J.U.; Zhang, S.; Lu, J.; Yang, L.; Ni, D.; Pang, X.; Lv, J. Identification of a novel type I pullulanase from Fervidobacterium nodosum Rt17-B1, with high thermostability and suitable optimal pH. Int. J. Biol. Macromol. 2020, 143, 424–433. [Google Scholar] [CrossRef]
- Chen, A.; Li, Y.; Nie, J.; McNeil, B.; Jeffrey, L.; Yang, Y.; Bai, Z. Protein engineering of Bacillus acidopullulyticus pullulanase for enhanced thermostability using in silico data driven rational design methods. Enzyme Microb. Technol. 2015, 78, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, K.-M.; Choi, K.-H.; Park, C.-S.; Kim, G.-E.; Kim, D.; Cha, J. Molecular cloning and biochemical characterization of a heat-stable type I pullulanase from Thermotoga neapolitana. Enzyme Microb. Technol. 2011, 48, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dong, F.; Lin, L.; He, D.; Wei, W.; Wei, D. N-terminal domain truncation and domain insertion-based engineering of a novel thermostable type i pullulanase from Geobacillus thermocatenulatus. J. Agric. Food Chem. 2018, 66, 10788–10798. [Google Scholar] [CrossRef]
- Lu, Z.; Hu, X.; Shen, P.; Wang, Q.; Zhou, Y.; Zhang, G.; Ma, Y. A pH-stable, detergent and chelator resistant type I pullulanase from Bacillus pseudofirmus 703 with high catalytic efficiency. Int. J. Biol. Macromol. 2018, 109, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-Y.; Guo, Z.-W.; Wu, X.-L.; Ou, X.-Y.; Zong, M.-H.; Lou, W.-Y. Recombinant expression and characterization of a novel cold-adapted type I pullulanase for efficient amylopectin hydrolysis. J. Biotechnol. 2020, 313, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ma, J.; Guo, S.; Wei, D.-Z. A type I pullulanase of Bacillus cereus Nws-bc5 screening from stinky tofu brine: Functional expression in Escherichia coli and Bacillus subtilis and enzyme characterization. Process Biochem. 2014, 49, 1893–1902. [Google Scholar] [CrossRef]
- Prakash, N.; Gupta, S.; Ansari, M.; Khan, Z.A.; Suneetha, V. Production of economically important products by the use of pullulanase enzyme. Int. J. Sci. Innovs. 2012, 2, 266–273. [Google Scholar]
- Birt, D.F.; Boylston, T.; Hendrich, S.; Jane, J.L.; Hollis, J.; Li, L.; McClelland, J.; Moore, S.; Phillips, G.J.; Rowling, M.; et al. Resistant starch: Promise for improving human health. Adv. Nutr. 2013, 4, 587–601. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Chen, Y.; Yu, S.; Gao, Q. Preparation and properties of RS III from waxy maize starch with pullulanase. Food Hydrocoll. 2013, 33, 19–25. [Google Scholar] [CrossRef]
- Tu, D.; Ou, Y.; Zheng, Y.; Zhang, Y.; Zheng, B.; Zeng, H. Effects of freeze-thaw treatment and pullulanase debranching on the structural properties and digestibility of lotus seed starch-glycerin monostearin complexes. Int. J. Biol. Macromol. 2021, 177, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Breuninger, W.F.; Piyachomkwan, K.; Sriroth, K. Chapter 12-Tapioca/cassava starch: Production and use. In Starch, 3rd ed.; BeMiller, J., Whistler, R., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 541–568. [Google Scholar]
- Prongjit, D.; Lekakarn, H.; Bunterngsook, B.; Aiewviriyasakul, K.; Sritusnee, W.; Champreda, V. Functional characterization of recombinant raw starch degrading α-amylase from Roseateles terrae HL11 and its application on cassava pulp saccharification. Catalysts 2022, 12, 647. [Google Scholar] [CrossRef]
- Callister, W.D.; Rethwisch, D.G. Materials Science and Engineering an Introduction, 10th ed.; Wiley: Hoboken, NJ, USA, 2018. [Google Scholar]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; I Hurwitz, D.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2001. [Google Scholar]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]









| Substrates | Relative Activity (%) | Specific Activity (U/mg Protein) |
|---|---|---|
| Pullulan | 100.00 ± 1.96 | 181.14 ± 3.55 |
| Amylopectin | 9.78 ± 0.50 | 17.706 ± 0.92 |
| Amylose | a ND | a ND |
| Soluble strach | 29.57 ± 0.78 | 53.19 ± 2.02 |
| Starch from cassava | 33.99 ± 0.34 | 61.57 ± 0.61 |
| Starch from rice | 29.12 ± 0.25 | 52.75 ± 0.45 |
| Starch from potato | 31.51 ± 1.39 | 57.07 ± 2.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prongjit, D.; Lekakarn, H.; Bunterngsook, B.; Aiewviriyasakul, K.; Sritusnee, W.; Arunrattanamook, N.; Champreda, V. In-Depth Characterization of Debranching Type I Pullulanase from Priestia koreensis HL12 as Potential Biocatalyst for Starch Saccharification and Modification. Catalysts 2022, 12, 1014. https://0-doi-org.brum.beds.ac.uk/10.3390/catal12091014
Prongjit D, Lekakarn H, Bunterngsook B, Aiewviriyasakul K, Sritusnee W, Arunrattanamook N, Champreda V. In-Depth Characterization of Debranching Type I Pullulanase from Priestia koreensis HL12 as Potential Biocatalyst for Starch Saccharification and Modification. Catalysts. 2022; 12(9):1014. https://0-doi-org.brum.beds.ac.uk/10.3390/catal12091014
Chicago/Turabian StyleProngjit, Daran, Hataikarn Lekakarn, Benjarat Bunterngsook, Katesuda Aiewviriyasakul, Wipawee Sritusnee, Nattapol Arunrattanamook, and Verawat Champreda. 2022. "In-Depth Characterization of Debranching Type I Pullulanase from Priestia koreensis HL12 as Potential Biocatalyst for Starch Saccharification and Modification" Catalysts 12, no. 9: 1014. https://0-doi-org.brum.beds.ac.uk/10.3390/catal12091014