A Theoretical and Experimental Study for Enzymatic Biodiesel Production from Babassu Oil (Orbignya sp.) Using Eversa Lipase
Abstract
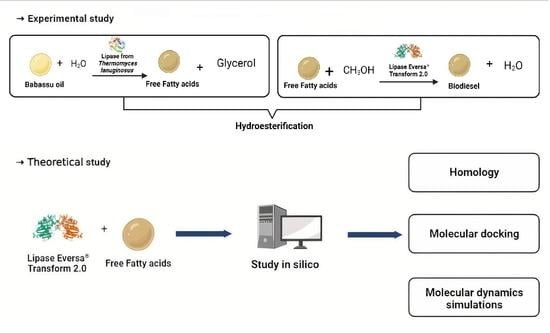
:1. Introduction
2. Results and Discussion
2.1. Enzymatic Hydrolysis
2.2. Taguchi Planning-Optimization of the Production of Methyl Esters of Babassu Fatty Acids
2.2.1. S/N Ratio Analysis
2.2.2. Analysis of Variance (ANOVA)
2.3. Physicochemical Characterization of the Oil Produced
2.4. Theoretical Study
2.4.1. Protein Modeling by Homology
2.4.2. Molecular Docking
2.4.3. Molecular Dynamics Simulations
Root Mean Square Deviation—RMSD
Hydrogen Bonds
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Hydroesterification
3.2.2. Gas Chromatography–Mass Spectrometry (GC/MS) Analysis
3.2.3. Experimental Design and Statistical Analysis (Taguchi Method)
3.2.4. Physicochemical Characterization of Babassu FAME
Kinematic Viscosity
3.2.5. In Silico Study
Protein Modeling by Homology
Molecular Docking
Molecular Dynamics Simulations
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Li, H.; Pan, H.; Liu, X.; Yang, K.; Huang, S.; Yang, S. Efficient Production of Biodiesel with Promising Fuel Properties from Koelreuteria Integrifoliola Oil Using a Magnetically Recyclable Acidic Ionic Liquid. Energy Convers. Manag. 2017, 138, 45–53. [Google Scholar] [CrossRef]
- Abudu, H.; Sai, R. Examining Prospects and Challenges of Ghana’s Petroleum Industry: A Systematic Review. Energy Rep. 2020, 6, 841–858. [Google Scholar] [CrossRef]
- Goh, B.H.H.; Ong, H.C.; Cheah, M.Y.; Chen, W.H.; Yu, K.L.; Mahlia, T.M.I. Sustainability of Direct Biodiesel Synthesis from Microalgae Biomass: A Critical Review. Renew. Sustain. Energy Rev. 2019, 107, 59–74. [Google Scholar] [CrossRef]
- Peng, L.; Fu, D.; Chu, H.; Wang, Z.; Qi, H. Biofuel Production from Microalgae: A Review. Environ. Chem. Lett. 2020, 18, 285–297. [Google Scholar] [CrossRef]
- Lima, P.J.M.; da Silva, R.M.; Neto, C.A.C.G.; Gomes e Silva, N.C.; Souza, J.E.d.S.; Nunes, Y.L.; Sousa dos Santos, J.C. An Overview on the Conversion of Glycerol to Value-Added Industrial Products via Chemical and Biochemical Routes. Biotechnol. Appl. Biochem. 2021. [Google Scholar] [CrossRef]
- Temóteo, R.L.; da Silva, M.J.; de Ávila Rodrigues, F.; da Silva, W.F.; de Jesus Silva, D.; Oliveira, C.M. A Kinetic Investigation of Triacetin Methanolysis and Assessment of the Stability of a Sulfated Zirconium Oxide Catalyst. J. Am. Oil Chem. Soc. 2018, 95, 865–874. [Google Scholar] [CrossRef]
- Staufenberg, G.; Graupner, N.; Müssig, J.; Tripathi, M.; Bhatnagar, A.; Pandey, K.K.; De, M.; Salgado, F.; Abioye, A.M.; Junoh, M.M.; et al. Preparation of Activated Carbon from Babassu Endocarpunder Microwave Radiation by Physical Activation. IOP Conf. Series Earth Environ. Sci. 2018, 105, 012116. [Google Scholar] [CrossRef] [Green Version]
- Sales, M.B.; Borges, P.T.; Ribeiro Filho, M.N.; Miranda da Silva, L.R.; Castro, A.P.; Sanders Lopes, A.A.; Chaves de Lima, R.K.; de Sousa Rios, M.A.; Santos, J.C.S. dos Sustainable Feedstocks and Challenges in Biodiesel Production: An Advanced Bibliometric Analysis. Bioengineering 2022, 9, 539. [Google Scholar] [CrossRef]
- Mota, G.F.; de Sousa, I.G.; de Oliveira, A.L.B.; Cavalcante, A.L.G.; Moreira, K.d.S.; Cavalcante, F.T.T.; Souza, J.E.D.S.; Falcão, R.d.A.; Rocha, T.G.; Valério, R.B.R.; et al. Biodiesel Production from Microalgae Using Lipase-Based Catalysts: Current Challenges and Prospects. Algal Res. 2022, 62, 102616. [Google Scholar] [CrossRef]
- Wancura, J.H.C.; Rosset, D.v.; Mazutti, M.A.; Ugalde, G.A.; de Oliveira, J.V.; Tres, M.v.; Jahn, S.L. Improving the Soluble Lipase–Catalyzed Biodiesel Production through a Two-Step Hydroesterification Reaction System. Appl. Microbiol. Biotechnol. 2019, 103, 7805–7817. [Google Scholar] [CrossRef]
- Cavalcante, F.T.T.; Cavalcante, A.L.G.; de Sousa, I.G.; Neto, F.S.; dos Santos, J.C.S. Current Status and Future Perspectives of Supports and Protocols for Enzyme Immobilization. Catalysts 2021, 11, 1222. [Google Scholar] [CrossRef]
- Monteiro, R.R.C.; dos Santos, J.C.S.; Alcántara, A.R.; Fernandez-Lafuente, R. Enzyme-Coated Micro-Crystals: An Almost Forgotten but Very Simple and Elegant Immobilization Strategy. Catalysts 2020, 10, 891. [Google Scholar] [CrossRef]
- de Oliveira, A.L.B.; Cavalcante, F.T.T.; Moreira, K.S.; Monteiro, R.R.C.; Rocha, T.G.; Souza, J.E.S.; da Fonseca, A.M.; Lopes, A.A.S.; Guimarães, A.P.; de Lima, R.K.C.; et al. Lipases Immobilized onto Nanomaterials as Biocatalysts in Biodiesel Production: Scientific Context, Challenges, and Opportunities. Rev. Virtual Química 2021, 13, 875–891. [Google Scholar] [CrossRef]
- Marques da Fonseca, A.; Bezerra de Freitas, Í.; Baltazar Soares, N.; Aurecio Morais de Araújo, F.; Menezes Gaieta, E.; Cleiton Sousa dos Santos, J.; Carlos Nogueira Sobrinho, A.; Silva Marinho, E.; Paulo Colares, R. Synthesis, Biological Activity, and In Silico Study of Bioesters Derived from Bixin by the CALB Enzyme. Biointerface Res. Appl. Chem. 2021, 12, 5901–5917. [Google Scholar] [CrossRef]
- Cavalcante, A.L.G.; Chaves, A.V.; Fechine, P.B.A.; Holanda Alexandre, J.Y.N.; Freire, T.M.; Davi, D.M.B.; Neto, F.S.; de Sousa, I.G.; da Silva Moreira, K.; de Oliveira, A.L.B.; et al. Chemical Modification of Clay Nanocomposites for the Improvement of the Catalytic Properties of Lipase A from Candida Antarctica. Process Biochem. 2022, 120, 1–14. [Google Scholar] [CrossRef]
- Bonazza, H.L.; Manzo, R.M.; dos Santos, J.C.S.; Mammarella, E.J. Operational and Thermal Stability Analysis of Thermomyces Lanuginosus Lipase Covalently Immobilized onto Modified Chitosan Supports. Appl. Biochem. Biotechnol. 2018, 184, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, R.R.C.; de Oliveira, A.L.B.; de Menezes, F.L.; de Souza, M.C.M.; Fechine, P.B.A.; dos Santos, J.C.S. Improvement of Enzymatic Activity and Stability of Lipase A from Candida Antartica onto Halloysite Nanotubes with Taguchi Method for Optimized Immobilization. Appl. Clay Sci. 2022, 228, 106634. [Google Scholar] [CrossRef]
- Silva, A.R.M.; Alexandre, J.Y.N.H.; Souza, J.E.S.; Neto, J.G.L.; De, S.; Júnior, P.G.; Rocha, M.V.P.; dos Santos, J.C.S.; Silva, A.R.M.; Alexandre, J.Y.N.H.; et al. The Chemistry and Applications of Metal–Organic Frameworks (MOFs) as Industrial Enzyme Immobilization Systems. Molecules 2022, 27, 4529. [Google Scholar] [CrossRef]
- Velasco-Lozano, S.; Rocha-Martin, J.; Santos, J.C.S. dos Editorial: Designing Carrier-Free Immobilized Enzymes for Biocatalysis. Front. Bioeng. Biotechnol. 2022, 10, 823. [Google Scholar] [CrossRef]
- Souza, J.E.D.S.; Oliveira, G.P.D.; Alexandre, J.Y.N.H.; Da, J.E.; Souza, S.; de Oliveira, G.P.; Alexandre, J.Y.N.H.; Neto, J.G.L.; Sales, M.B.; De, P.G.; et al. A Comprehensive Review on the Use of Metal–Organic Frameworks (MOFs) Coupled with Enzymes as Biosensors. Electrochem 2022, 3, 89–113. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Barbosa, O.; Hernandez, K.; Santos, J.; Rodrigues, R.; Fernandez-Lafuente, R. Evaluation of Styrene-Divinylbenzene Beads as a Support to Immobilize Lipases. Molecules 2014, 19, 7629–7645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, W.C.A.; Luiz, J.H.H.; Fernandez-Lafuente, R.; Hirata, D.B.; Mendes, A.A. Eco-Friendly Production of Trimethylolpropane Triesters from Refined and Used Soybean Cooking Oils Using an Immobilized Low-Cost Lipase (Eversa>® Transform 2.0) as Heterogeneous Catalyst. Biomass Bioenergy 2021, 155, 106302. [Google Scholar] [CrossRef]
- Chang, M.Y.; Chan, E.S.; Song, C.P. Biodiesel Production Catalysed by Low-Cost Liquid Enzyme Eversa® Transform 2.0: Effect of Free Fatty Acid Content on Lipase Methanol Tolerance and Kinetic Model. Fuel 2021, 283, 119266. [Google Scholar] [CrossRef]
- Cavalcante, F.T.T.; da Fonseca, A.M.; Holanda Alexandre, J.Y.N.; dos Santos, J.C.S. A Stepwise Docking and Molecular Dynamics Approach for Enzymatic Biolubricant Production Using Lipase Eversa® Transform as a Biocatalyst. Ind. Crop. Prod. 2022, 187, 115450. [Google Scholar] [CrossRef]
- Qin, X.; Zhong, J.; Wang, Y. A Mutant T1 Lipase Homology Modeling, and Its Molecular Docking and Molecular Dynamics Simulation with Fatty Acids. J. Biotechnol. 2021, 337, 24–34. [Google Scholar] [CrossRef]
- Moreira, K.d.S.; de Oliveira, A.L.B.; Júnior, L.S.d.M.; Monteiro, R.R.C.; da Rocha, T.N.; Menezes, F.L.; Fechine, L.M.U.D.; Denardin, J.C.; Michea, S.; Freire, R.M.; et al. Lipase From Rhizomucor Miehei Immobilized on Magnetic Nanoparticles: Performance in Fatty Acid Ethyl Ester (FAEE) Optimized Production by the Taguchi Method. Front. Bioeng. Biotechnol. 2020, 8, 693. [Google Scholar] [CrossRef]
- Souza, J.E.S.; Monteiro, R.R.C.; Rocha, T.G.; Moreira, K.S.; Cavalcante, F.T.T.; de Sousa Braz, A.K.; de Souza, M.C.M.; dos Santos, J.C.S. Sonohydrolysis Using an Enzymatic Cocktail in the Preparation of Free Fatty Acid. 3 Biotech 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Mohd Hussin, F.N.N.; Attan, N.; Wahab, R.A. Taguchi Design-Assisted Immobilization of Candida Rugosa Lipase onto a Ternary Alginate/Nanocellulose/Montmorillonite Composite: Physicochemical Characterization, Thermal Stability and Reusability Studies. Enzym. Microb. Technol. 2020, 136, 109506. [Google Scholar] [CrossRef]
- Li, S.; Zhong, L.; Wang, H.; Li, J.; Cheng, H.; Ma, Q. Process Optimization of Polyphenol Oxidase Immobilization: Isotherm, Kinetic, Thermodynamic and Removal of Phenolic Compounds. Int. J. Biol. Macromol. 2021, 185, 792–803. [Google Scholar] [CrossRef]
- Ameri, A.; Shakibaie, M.; Khoobi, M.; Faramarzi, M.A.; Ameri, A.; Forootanfar, H. Immobilization of Thermoalkalophilic Lipase from Bacillus Atrophaeus FSHM2 on Amine-Modified Graphene Oxide Nanostructures: Statistical Optimization and Its Application for Pentyl Valerate Synthesis. Appl. Biochem. Biotechnol. 2020, 191, 579–604. [Google Scholar] [CrossRef]
- Babaki, M.; Yousefi, M.; Habibi, Z.; Mohammadi, M. Process Optimization for Biodiesel Production from Waste Cooking Oil Using Multi-Enzyme Systems through Response Surface Methodology. Renew. Energy 2017, 105, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Arana-Peña, S.; Carballares, D.; Morellon-Sterlling, R.; Berenguer-Murcia, Á.; Alcántara, A.R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Enzyme Co-Immobilization: Always the Biocatalyst Designers’ Choice…or Not? Biotechnol. Adv. 2021, 51, 107584. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, R.R.C.; Arana-Peña, S.; da Rocha, T.N.; Miranda, L.P.; Berenguer-Murcia, Á.; Tardioli, P.W.; dos Santos, J.C.S.; Fernandez-Lafuente, R. Liquid Lipase Preparations Designed for Industrial Production of Biodiesel. Is It Really an Optimal Solution? Renew. Energy 2021, 164, 1566–1587. [Google Scholar] [CrossRef]
- Sun, S.; Guo, J.; Chen, X. Biodiesel Preparation from Semen Abutili (Abutilon Theophrasti Medic.) Seed Oil Using Low-Cost Liquid Lipase Eversa® Transform 2.0 as a Catalyst. Ind. Crop. Prod. 2021, 169, 113643. [Google Scholar] [CrossRef]
- Adewale, P.; Vithanage, L.N.; Christopher, L. Optimization of Enzyme-Catalyzed Biodiesel Production from Crude Tall Oil Using Taguchi Method. Energy Convers. Manag. 2017, 154, 81–91. [Google Scholar] [CrossRef]
- Knothe, G.; Steidley, K.R. Kinematic Viscosity of Biodiesel Fuel Components and Related Compounds. Influence of Compound Structure and Comparison to Petrodiesel Fuel Components. Fuel 2005, 84, 1059–1065. [Google Scholar] [CrossRef]
- Ramírez Verduzco, L.F. Density and Viscosity of Biodiesel as a Function of Temperature: Empirical Models. Renew. Sustain. Energy Rev. 2013, 19, 652–665. [Google Scholar] [CrossRef]
- Tam, B.; Sinha, S.; Wang, S.M. Combining Ramachandran Plot and Molecular Dynamics Simulation for Structural-Based Variant Classification: Using TP53 Variants as Model. Comput. Struct. Biotechnol. J. 2020, 18, 4033–4039. [Google Scholar] [CrossRef]
- Sarkar, S.; Banerjee, A.; Chakraborty, N.; Soren, K.; Chakraborty, P.; Bandopadhyay, R. Structural-Functional Analyses of Textile Dye Degrading Azoreductase, Laccase and Peroxidase: A Comparative in Silico Study. Electron. J. Biotechnol. 2020, 43, 48–54. [Google Scholar] [CrossRef]
- Bronowska, A.K. Thermodynamics of Ligand-Protein Interactions: Implications for Molecular Design. Thermodyn. Interact. Stud. Solids Liq. Gases 2011. [Google Scholar] [CrossRef]
- Lan, D.; Zhao, G.; Holzmann, N.; Yuan, S.; Wang, J.; Wang, Y. Structure-Guided Rational Design of a Mono- And Diacylglycerol Lipase from Aspergillus Oryzae: A Single Residue Mutant Increases the Hydrolysis Ability. J. Agric. Food Chem. 2021, 69, 5344–5352. [Google Scholar] [CrossRef]
- Zhang, M.; Li, Q.; Lan, X.; Li, X.; Zhang, Y.; Wang, Z.; Zheng, J. Directed Evolution of Aspergillus Oryzae Lipase for the Efficient Resolution of (R,S)-Ethyl-2-(4-Hydroxyphenoxy) Propanoate. Bioprocess Biosyst. Eng. 2020, 43, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, J.; Kodali, S.; Balle, T.; Chen, B.; Guo, Z. Liquid Lipases for Enzymatic Concentration of N-3 Polyunsaturated Fatty Acids in Monoacylglycerols via Ethanolysis: Catalytic Specificity and Parameterization. Bioresour Technol 2017, 224, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Tamboli, A.S.; Waghmare, P.R.; Khandare, R.v.; Govindwar, S.P. Comparative Analyses of Enzymatic Activity, Structural Study and Docking of Fungal Cellulases. Gene Rep. 2017, 9, 54–60. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Y.; Zeng, G.; Shao, B.; Chen, M.; Li, Z.; Jiang, Y.; Liu, Y.; Zhang, Y.; Zhong, H. Application of Molecular Docking for the Degradation of Organic Pollutants in the Environmental Remediation: A Review. Chemosphere 2018, 203, 139–150. [Google Scholar] [CrossRef]
- Fatma, T.; Zafar, Z.; Fatima, S.; Paracha, R.Z.; Adnan, F.; Sheikh, Z.; Virk, N.; Bhatti, M.F. Computational Assessment of Botrytis Cinerea Lipase for Biofuel Production. Catalysts 2021, 11, 1319. [Google Scholar] [CrossRef]
- Williams, D.H.; Westwell, M.S. Aspects of Weak Interactions. Chem. Soc. Rev. 1998, 27, 57–64. [Google Scholar] [CrossRef]
- Guimarães, J.R.; Miranda, L.P.; Fernandez-Lafuente, R.; Tardioli, P.W. Immobilization of Eversa® Transform via CLEA Technology Converts It in a Suitable Biocatalyst for Biolubricant Production Using Waste Cooking Oil. Molecules 2021, 26, 193. [Google Scholar] [CrossRef]
- Joshi, T.; Joshi, T.; Sharma, P.; Chandra, S.; Pande, V. Molecular Docking and Molecular Dynamics Simulation Approach to Screen Natural Compounds for Inhibition of Xanthomonas Oryzae Pv. Oryzae by Targeting Peptide Deformylase. J. Biomol. Struct. Dyn. 2020, 39, 823–840. [Google Scholar] [CrossRef]
- Ragunathan, A.; Malathi, K.; Anbarasu, A. MurB as a Target in an Alternative Approach to Tackle the Vibrio Cholerae Resistance Using Molecular Docking and Simulation Study. J. Cell. Biochem. 2018, 119, 1726–1732. [Google Scholar] [CrossRef]
- Qu, P.; Li, D.; Lazim, R.; Xu, R.; Xiao, D.; Wang, F.; Li, X.; Zhang, Y. Improved Thermostability of Thermomyces Lanuginosus Lipase by Molecular Dynamics Simulation and in Silico Mutation Prediction and Its Application in Biodiesel Production. Fuel 2022, 327, 125039. [Google Scholar] [CrossRef]
- Moreira, K.S.; Moura, L.S.; Monteiro, R.R.C.; de Oliveira, A.L.B.; Valle, C.P.; Freire, T.M.; Fechine, P.B.A.; de Souza, M.C.M.; Fernandez-Lorente, G.; Guisan, J.M.; et al. Optimization of the Production of Enzymatic Biodiesel from Residual Babassu Oil (Orbignya Sp.) via RSM. Catalysts 2020, 10, 414. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, R.R.C.; Neto, D.M.A.; Fechine, P.B.A.; Lopes, A.A.S.; Gonçalves, L.R.B.; dos Santos, J.C.S.; de Souza, M.C.M.; Fernandez-Lafuente, R. Ethyl Butyrate Synthesis Catalyzed by Lipases A and B from Candida Antarctica Immobilized onto Magnetic Nanoparticles. Improvement of Biocatalysts’ Performance under Ultrasonic Irradiation. Int. J. Mol. Sci. 2019, 20, 5807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, T.G.; de L. Gomes, P.H.; de Souza, M.C.M.; Monteiro, R.R.C.; dos Santos, J.C.S. Lipase Cocktail for Optimized Biodiesel Production of Free Fatty Acids from Residual Chicken Oil. Catal Lett. 2021, 151, 1155–1166. [Google Scholar] [CrossRef]
- Figueredo, I.D.M.; Rios, M.A.D.S.; Cavalcante, C.L.; Luna, F.M.T. Effects of Amine and Phenolic Based Antioxidants on the Stability of Babassu Biodiesel Using Rancimat and Differential Scanning Calorimetry Techniques. Ind. Eng. Chem. Res. 2020, 59, 18–24. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- BSI BS EN 14103; Fat and Oil Derivatives-Fatty Acid Methyl Esters (FAME)-Determination of Ester and Linolenic Acid Methyl Ester Contents. European Committee for Standardization, Management Centre: Bruxelles, Belgium, 2011; Volume 17, pp. 1–22.
- Moreira, K.D.S.; de Oliveira, A.L.B.; Júnior, L.S.D.M.; de Sousa, I.G.; Cavalcante, A.L.G.; Neto, F.S.; Valério, R.B.R.; Chaves, A.V.; Fonseca, T.D.S.; Cruz, D.M.V.; et al. Taguchi Design-Assisted Co-Immobilization of Lipase A and B from Candida Antarctica onto Chitosan: Characterization, Kinetic Resolution Application, and Docking Studies. Chem. Eng. Res. Des. 2022, 177, 223–244. [Google Scholar] [CrossRef]
- Chakraborty, R.; RoyChowdhury, D. Fish Bone Derived Natural Hydroxyapatite-Supported Copper Acid Catalyst: Taguchi Optimization of Semibatch Oleic Acid Esterification. Chem. Eng. J. 2013, 215–216, 491–499. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Bedoya, O.F.; Tischer, I. Detección de Homología Remota de Proteínas Usando Modelos 3D Enriquecidos Con Propiedades Fisicoquímicas. Ing. Y Compet. 2015, 17, 75–84. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Cheatham, T.E.; Miller, J.L.; Fox, T.; Darden, T.A.; Kollman, P.A. Molecular Dynamics Simulations on Solvated Biomolecular Systems: The Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, RNA, and Proteins. J. Am. Chem. Soc. 1995, 117, 4193–4194. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.v.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Field, M.J.; Albe, M.; Bret, C.; Proust-De Martin, F.; Thomas, A. The Dynamo Library for Molecular Simulations Using Hybrid Quantum Mechanical and Molecular Mechanical Potentials. J. Comput. Chem. 2000, 21, 1088–1100. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]







| Reaction | Time (h) | Temperature (°C) | Molar Ratio (FFA/Methanol) | Biocatalyst (% w/w) | Conversion (%) | S/N |
|---|---|---|---|---|---|---|
| 1 | 1 | 25 | 1:1 | 0.1 | 3.16 ± 0.48 | 9.99 |
| 2 | 1 | 40 | 1:5 | 0.5 | 58.79 ± 0.078 | 35.39 |
| 3 | 1 | 55 | 1:9 | 0.9 | 19.43 ± 0.66 | 25.77 |
| 4 | 3 | 25 | 1:5 | 0.9 | 95.20 ± 0.031 | 39.57 |
| 5 | 3 | 40 | 1:9 | 0.1 | 7.75 ± 0.30 | 17.79 |
| 6 | 3 | 55 | 1:1 | 0.5 | 8.44 ± 0.53 | 18.52 |
| 7 | 5 | 25 | 1:9 | 0.5 | 94.65 ± 0.036 | 39.52 |
| 8 | 5 | 40 | 1:1 | 0.9 | 75.36 ± 0.51 | 37.54 |
| 9 | 5 | 55 | 1:5 | 0.1 | 2.80 ± 0.18 | 8.94 |
| Factors/Levels | Time (h) | Temperature (°C) | The Molar Ratio (FFA/Methanol) | Biocatalyst (% w/w) |
|---|---|---|---|---|
| 1 | 23.72 | 29.70 | 22.02 | 12.24 |
| 2 | 25.29 | 30.24 | 27.97 | 31.14 |
| 3 | 28.67 | 17.74 | 27.69 | 34.29 |
| Delta | 4.95 | 12.50 | 5.95 | 22.05 |
| Ranking | 4 | 2 | 3 | 1 |
| Factors | SS | DF | MS | F-value | p-Value | Contribution (%) |
|---|---|---|---|---|---|---|
| Time | 38.40 | 2 | - | - | - | 3.05% |
| Temperature | 299.30 | 2 | 149.65 | 7.79 | 0.11 | 23.77% |
| Molar ratio | 67.62 | 2 | 33.812 | 1.76 | 0.36 | 5.37% |
| Biocatalyst | 853.63 | 2 | 426.817 | 22.23 | 0.043 | 67.80% |
| Residual | 38.40 | 2 | 19.20 | - | ||
| Total | 1258.98 | 6 | - | - | - |
| Substrate | Chosen Pose | Binding Affinity (kCal/mol) | Vina RMSD (Å) |
|---|---|---|---|
| Octanoic acid | 2 | −5.1 | 1.566 |
| Decanoic acid | 2 | −5.4 | 1.561 |
| Dodecanoic acid | 4 | −5.6 | 1.658 |
| Tetradecanoic acid | 2 | −5.8 | 0.970 |
| Hexadecanoic acid | 4 | −5.8 | 2.296 |
| cis-9-octadecenoic acid | 5 | −6.2 | 1.426 |
| Substrate | Waste Involved | ||
|---|---|---|---|
| Hydrogen Bonds | Van der Waals Interactions | Hydrophobic Interactions | |
| Octanoic acid | - | ASP276, HIS268, LEU154, SER91, SER153, TYR29 | ILE94, PHE265, TYR92, |
| Decanoic acid | TYR92 (2.94 Å) | ASP276, HIS268 LEU154, SER91, SER153, TYR29 | ILE94, PHE265 |
| Dodecanoic acid | HIS152 (3.06 Å) | ASP276, HIS268, LEU154, SER153, TYR29 | ILE94, LEU262, PHE265, TYR92 |
| Tetradecanoic acid | - | ASP276, HIS266, HIS268, HIS274, ILE94, LEU154, LEU285, SER153, TYR29 | LEU283, PHE265, TYR92, VAL269 |
| Hexadecanoic acid | HIS152 (2.73 Å), HIS268 (2.96 Å), SER153 (2.86 Å), TYR29 (2.78 Å), TYR29 (2.59 Å) | ASP276, LEU154, LEU262, VAL269 | ILE94, LEU283, PHE265, TYR92 |
| cis-9-octadecenoic acid | SER 91 (2.27 Å) | APS276, HIS266, HIS268, HIS274, LEU154, TYR29, SER 153, SER93 | ILE94, LEU283, PHE265, VAL269, TYR92 |
| Time (hours) | Temperature (°C) | Molar Ratio (FFA/Methanol) | Biocatalyst (% w/w) |
|---|---|---|---|
| Level 1 (L1) | 1 | 25 | 0.1 |
| Level 2 (L2) | 3 | 40 | 0.5 |
| Level 3 (L3) | 5 | 55 | 0.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandre, J.Y.N.H.; Cavalcante, F.T.T.; Freitas, L.M.; Castro, A.P.; Borges, P.T.; de Sousa Junior, P.G.; Filho, M.N.R.; Lopes, A.A.S.; da Fonseca, A.M.; Lomonaco, D.; et al. A Theoretical and Experimental Study for Enzymatic Biodiesel Production from Babassu Oil (Orbignya sp.) Using Eversa Lipase. Catalysts 2022, 12, 1322. https://0-doi-org.brum.beds.ac.uk/10.3390/catal12111322
Alexandre JYNH, Cavalcante FTT, Freitas LM, Castro AP, Borges PT, de Sousa Junior PG, Filho MNR, Lopes AAS, da Fonseca AM, Lomonaco D, et al. A Theoretical and Experimental Study for Enzymatic Biodiesel Production from Babassu Oil (Orbignya sp.) Using Eversa Lipase. Catalysts. 2022; 12(11):1322. https://0-doi-org.brum.beds.ac.uk/10.3390/catal12111322
Chicago/Turabian StyleAlexandre, Jeferson Yves Nunes Holanda, Francisco Thálysson Tavares Cavalcante, Lara Matias Freitas, Alyne Prudêncio Castro, Pedro Tavares Borges, Paulo Gonçalves de Sousa Junior, Manoel Nazareno Ribeiro Filho, Ada Amelia Sanders Lopes, Aluisio Marques da Fonseca, Diego Lomonaco, and et al. 2022. "A Theoretical and Experimental Study for Enzymatic Biodiesel Production from Babassu Oil (Orbignya sp.) Using Eversa Lipase" Catalysts 12, no. 11: 1322. https://0-doi-org.brum.beds.ac.uk/10.3390/catal12111322