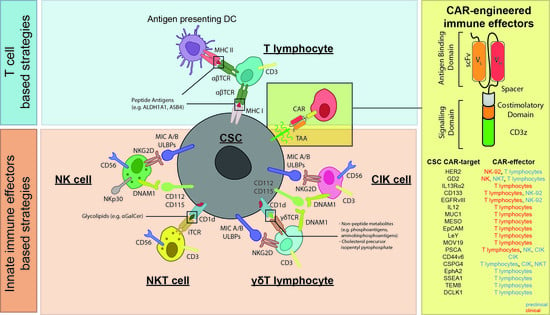
Cellular Immunotherapy Targeting Cancer Stem Cells: Preclinical Evidence and Clinical Perspective
Abstract
:1. Introduction
2. Identification and Immunological Properties of CSCs
2.1. Immunological Features
2.2. Signaling Pathways’ Alterations in Cancer Stem Cells
2.3. CSCs Markers and Tumor Associated Antigens (TAA)
3. Cellular Immunotherapy Targeting CSCs
3.1. T Cell-Based Strategies Targeting CSC
3.2. Innate Immune Effectors Targeting CSCs
3.2.1. NK Cells
3.2.2. CIK and NKT Cells
3.2.3. γδ T Cells
4. Conclusions and Challenging Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef]
- Cojoc, M.; Mäbert, K.; Muders, M.H.; Dubrovska, A. A role for cancer stem cells in therapy resistance: Cellular and molecular mechanisms. Semin. Cancer Biol. 2015, 31, 16–27. [Google Scholar] [CrossRef]
- Tang, D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef]
- Hirschmann-Jax, C.; Foster, A.E.; Wulf, G.G.; Nuchtern, J.G.; Jax, T.W.; Gobel, U.; Goodell, M.A.; Brenner, M.K. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 14228–14233. [Google Scholar] [CrossRef] [Green Version]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell 2012, 10, 717–728. [Google Scholar] [CrossRef] [Green Version]
- Quintana, E.; Shackleton, M.; Foster, H.R.; Fullen, D.R.; Sabel, M.S.; Johnson, T.M.; Morrison, S.J. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell 2010, 18, 510–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntly, B.J.; Gilliland, D.G. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat. Rev. Cancer 2005, 5, 311–321. [Google Scholar] [CrossRef]
- Goodell, M.A.; Brose, K.; Paradis, G.; Conner, A.S.; Mulligan, R.C. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 1996, 183, 1797–1806. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- van den Hoogen, C.; van der Horst, G.; Cheung, H.; Buijs, J.T.; Lippitt, J.M.; Guzmán-Ramírez, N.; Hamdy, F.C.; Eaton, C.L.; Thalmann, G.N.; Cecchini, M.G.; et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010, 70, 5163–5173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E.; et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Walcher, L.; Kistenmacher, A.K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef] [PubMed]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaggupilli, A.; Elkord, E. Significance of CD44 and CD24 as cancer stem cell markers: An enduring ambiguity. Clin. Dev. Immunol. 2012, 2012, 708036. [Google Scholar] [CrossRef] [Green Version]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Hong, I.S.; Jang, G.B.; Lee, H.Y.; Nam, J.S. Targeting cancer stem cells by using the nanoparticles. Int. J. Nanomed. 2015, 10, 251–260. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, J. The cancer stem cell gamble. Science 2015, 347, 226–229. [Google Scholar] [CrossRef]
- Kaur, G.; Sharma, P.; Dogra, N.; Singh, S. Eradicating Cancer Stem Cells: Concepts, Issues, and Challenges. Curr. Treat. Options Oncol. 2018, 19, 20. [Google Scholar] [CrossRef]
- Vlashi, E.; Pajonk, F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin. Cancer Biol. 2015, 31, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [Green Version]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Gardelli, C.; Russo, L.; Cipolla, L.; Moro, M.; Andriani, F.; Rondinone, O.; Nicotra, F.; Sozzi, G.; Bertolini, G.; Roz, L. Differential glycosylation of collagen modulates lung cancer stem cell subsets through β1 integrin-mediated interactions. Cancer Sci. 2020. [Google Scholar] [CrossRef]
- Dragu, D.L.; Necula, L.G.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Therapies targeting cancer stem cells: Current trends and future challenges. World J. Stem Cells 2015, 7, 1185–1201. [Google Scholar] [CrossRef]
- Leuci, V.; Casucci, G.M.; Grignani, G.; Rotolo, R.; Rossotti, U.; Vigna, E.; Gammaitoni, L.; Mesiano, G.; Fiorino, E.; Donini, C.; et al. CD44v6 as innovative sarcoma target for CAR-redirected CIK cells. Oncoimmunology 2018, 7, e1423167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leuci, V.; Donini, C.; Grignani, G.; Rotolo, R.; Mesiano, G.; Fiorino, E.; Gammaitoni, L.; D’Ambrosio, L.; Merlini, A.; Landoni, E.; et al. CSPG4-Specific CAR.CIK Lymphocytes as a Novel Therapy for the Treatment of Multiple Soft-Tissue Sarcoma Histotypes. Clin. Cancer Res. 2020. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, G.; Yuan, X.; Xu, M.; Wang, H.; Ji, J.; Konda, B.; Black, K.L.; Yu, J.S. Antigen-specific T-cell response from dendritic cell vaccination using cancer stem-like cell-associated antigens. Stem Cells 2009, 27, 1734–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Tang, D.G.; Rycaj, K. Cancer stem cells: Regulation programs, immunological properties and immunotherapy. Semin Cancer Biol. 2018, 52, 94–106. [Google Scholar] [CrossRef]
- Agudo, J.; Park, E.S.; Rose, S.A.; Alibo, E.; Sweeney, R.; Dhainaut, M.; Kobayashi, K.S.; Sachidanandam, R.; Baccarini, A.; Merad, M.; et al. Quiescent Tissue Stem Cells Evade Immune Surveillance. Immunity 2018, 48, 271–285.e275. [Google Scholar] [CrossRef] [Green Version]
- Di Tomaso, T.; Mazzoleni, S.; Wang, E.; Sovena, G.; Clavenna, D.; Franzin, A.; Mortini, P.; Ferrone, S.; Doglioni, C.; Marincola, F.M.; et al. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin. Cancer Res. 2010, 16, 800–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravindran, S.; Rasool, S.; Maccalli, C. The Cross Talk between Cancer Stem Cells/Cancer Initiating Cells and Tumor Microenvironment: The Missing Piece of the Puzzle for the Efficient Targeting of these Cells with Immunotherapy. Cancer Microenviron. 2019, 12, 133–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schatton, T.; Schütte, U.; Frank, N.Y.; Zhan, Q.; Hoerning, A.; Robles, S.C.; Zhou, J.; Hodi, F.S.; Spagnoli, G.C.; Murphy, G.F.; et al. Modulation of T-cell activation by malignant melanoma initiating cells. Cancer Res. 2010, 70, 697–708. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wang, H.; Wang, X.; Zhao, C. CD44, a marker of cancer stem cells, is positively correlated with PD-L1 expression and immune cells infiltration in lung adenocarcinoma. Cancer Cell Int. 2020, 20, 583. [Google Scholar] [CrossRef]
- Zhang, D.; Park, D.; Zhong, Y.; Lu, Y.; Rycaj, K.; Gong, S.; Chen, X.; Liu, X.; Chao, H.P.; Whitney, P.; et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat. Commun. 2016, 7, 10798. [Google Scholar] [CrossRef] [Green Version]
- Domingo-Domenech, J.; Vidal, S.J.; Rodriguez-Bravo, V.; Castillo-Martin, M.; Quinn, S.A.; Rodriguez-Barrueco, R.; Bonal, D.M.; Charytonowicz, E.; Gladoun, N.; de la Iglesia-Vicente, J.; et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell 2012, 22, 373–388. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Barr, J.; Kong, L.Y.; Wang, Y.; Wu, A.; Sharma, A.K.; Gumin, J.; Henry, V.; Colman, H.; Priebe, W.; et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol. Cancer Ther. 2010, 9, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Maccalli, C.; Parmiani, G.; Ferrone, S. Immunomodulating and Immunoresistance Properties of Cancer-Initiating Cells: Implications for the Clinical Success of Immunotherapy. Immunol. Investig. 2017, 46, 221–238. [Google Scholar] [CrossRef]
- Yoshimura, A.; Muto, G. TGF-β function in immune suppression. Curr. Top. Microbiol. Immunol. 2011, 350, 127–147. [Google Scholar] [CrossRef]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Fakhrejahani, F.; Tomita, Y.; Maj-Hes, A.; Trepel, J.B.; De Santis, M.; Apolo, A.B. Immunotherapies for bladder cancer: A new hope. Curr. Opin. Urol. 2015, 25, 586–596. [Google Scholar] [CrossRef]
- Erin, N.; Podnos, A.; Tanriover, G.; Duymuş, Ö.; Cote, E.; Khatri, I.; Gorczynski, R.M. Bidirectional effect of CD200 on breast cancer development and metastasis, with ultimate outcome determined by tumor aggressiveness and a cancer-induced inflammatory response. Oncogene 2015, 34, 3860–3870. [Google Scholar] [CrossRef]
- Kawasaki, B.T.; Farrar, W.L. Cancer stem cells, CD200 and immunoevasion. Trends Immunol. 2008, 29, 464–468. [Google Scholar] [CrossRef]
- Messai, Y.; Gad, S.; Noman, M.Z.; Le Teuff, G.; Couve, S.; Janji, B.; Kammerer, S.F.; Rioux-Leclerc, N.; Hasmim, M.; Ferlicot, S.; et al. Renal Cell Carcinoma Programmed Death-ligand 1, a New Direct Target of Hypoxia-inducible Factor-2 Alpha, is Regulated by von Hippel-Lindau Gene Mutation Status. Eur. Urol. 2016, 70, 623–632. [Google Scholar] [CrossRef]
- Lee, Y.; Shin, J.H.; Longmire, M.; Wang, H.; Kohrt, H.E.; Chang, H.Y.; Sunwoo, J.B. CD44+ Cells in Head and Neck Squamous Cell Carcinoma Suppress T-Cell-Mediated Immunity by Selective Constitutive and Inducible Expression of PD-L1. Clin. Cancer Res. 2016, 22, 3571–3581. [Google Scholar] [CrossRef] [Green Version]
- Zhi, Y.; Mou, Z.; Chen, J.; He, Y.; Dong, H.; Fu, X.; Wu, Y. B7H1 Expression and Epithelial-To-Mesenchymal Transition Phenotypes on Colorectal Cancer Stem-Like Cells. PLoS ONE 2015, 10, e0135528. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, K.E.; Zhao, E.; Li, W.; Shi, L.; Xie, G.; Jiang, B.; Wang, Y.; Li, R.; Zhang, P.; et al. B7-H1 enhances proliferation ability of gastric cancer stem-like cells as a receptor. Oncol. Lett. 2015, 9, 1833–1838. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Wang, C.Y. Targeting cancer stem cells in squamous cell carcinoma. Precis Clin. Med. 2019, 2, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Regenbrecht, C.R.; Lehrach, H.; Adjaye, J. Stemming cancer: Functional genomics of cancer stem cells in solid tumors. Stem Cell Rev. 2008, 4, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, O.K. Cancer stem cell genomics: The quest for early markers of malignant progression. Expert Rev. Mol. Diagn. 2009, 9, 545–554. [Google Scholar] [CrossRef]
- Karamboulas, C.; Ailles, L. Developmental signaling pathways in cancer stem cells of solid tumors. Biochim. Biophys. Acta 2013, 1830, 2481–2495. [Google Scholar] [CrossRef]
- Pasca di Magliano, M.; Hebrok, M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 2003, 3, 903–911. [Google Scholar] [CrossRef]
- Ingham, P.W.; Placzek, M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat. Rev. Genet. 2006, 7, 841–850. [Google Scholar] [CrossRef]
- Miele, L.; Miao, H.; Nickoloff, B.J. NOTCH signaling as a novel cancer therapeutic target. Curr. Cancer Drug. Targets 2006, 6, 313–323. [Google Scholar] [CrossRef]
- Crompton, T.; Outram, S.V.; Hager-Theodorides, A.L. Sonic hedgehog signalling in T-cell development and activation. Nat. Rev. Immunol. 2007, 7, 726–735. [Google Scholar] [CrossRef]
- de la Roche, M.; Ritter, A.T.; Angus, K.L.; Dinsmore, C.; Earnshaw, C.H.; Reiter, J.F.; Griffiths, G.M. Hedgehog signaling controls T cell killing at the immunological synapse. Science 2013, 342, 1247–1250. [Google Scholar] [CrossRef] [Green Version]
- Hirohashi, Y.; Torigoe, T.; Tsukahara, T.; Kanaseki, T.; Kochin, V.; Sato, N. Immune responses to human cancer stem-like cells/cancer-initiating cells. Cancer Sci. 2016, 107, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Michel, K.D.; Uhmann, A.; Dressel, R.; van den Brandt, J.; Hahn, H.; Reichardt, H.M. The hedgehog receptor patched1 in T cells is dispensable for adaptive immunity in mice. PLoS ONE 2013, 8, e61034. [Google Scholar] [CrossRef] [Green Version]
- Xie, J. The hedgehog’s trick for escaping immunosurveillance: The molecular mechanisms driving myeloid-derived suppressor cell recruitment in hedgehog signaling-dependent tumors. Oncoimmunology 2014, 3, e29180. [Google Scholar] [CrossRef] [Green Version]
- Jinushi, M.; Chiba, S.; Yoshiyama, H.; Masutomi, K.; Kinoshita, I.; Dosaka-Akita, H.; Yagita, H.; Takaoka, A.; Tahara, H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12425–12430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malladi, S.; Macalinao, D.G.; Jin, X.; He, L.; Basnet, H.; Zou, Y.; de Stanchina, E.; Massagué, J. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 2016, 165, 45–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casey, S.C.; Tong, L.; Li, Y.; Do, R.; Walz, S.; Fitzgerald, K.N.; Gouw, A.M.; Baylot, V.; Gütgemann, I.; Eilers, M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352, 227–231. [Google Scholar] [CrossRef] [Green Version]
- George, S.; Miao, D.; Demetri, G.D.; Adeegbe, D.; Rodig, S.J.; Shukla, S.; Lipschitz, M.; Amin-Mansour, A.; Raut, C.P.; Carter, S.L.; et al. Loss of PTEN Is Associated with Resistance to Anti-PD-1 Checkpoint Blockade Therapy in Metastatic Uterine Leiomyosarcoma. Immunity 2017, 46, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Cho, H.; Hong, S.O.; Oh, S.J.; Lee, H.J.; Cho, E.; Woo, S.R.; Song, J.S.; Chung, J.Y.; Son, S.W.; et al. LC3B upregulation by NANOG promotes immune resistance and stem-like property through hyperactivation of EGFR signaling in immune-refractory tumor cells. Autophagy 2020, 1–20. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Maccalli, C.; Volontè, A.; Cimminiello, C.; Parmiani, G. Immunology of cancer stem cells in solid tumours. Rev. Eur. J. Cancer 2014, 50, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Dannull, J.; Diener, P.A.; Prikler, L.; Fürstenberger, G.; Cerny, T.; Schmid, U.; Ackermann, D.K.; Groettrup, M. Prostate stem cell antigen is a promising candidate for immunotherapy of advanced prostate cancer. Cancer Res. 2000, 60, 5522–5528. [Google Scholar]
- Kiessling, A.; Schmitz, M.; Stevanovic, S.; Weigle, B.; Hölig, K.; Füssel, M.; Füssel, S.; Meye, A.; Wirth, M.P.; Rieber, E.P. Prostate stem cell antigen: Identification of immunogenic peptides and assessment of reactive CD8+ T cells in prostate cancer patients. Int. J. Cancer 2002, 102, 390–397. [Google Scholar] [CrossRef]
- Matsueda, S.; Kobayashi, K.; Nonaka, Y.; Noguchi, M.; Itoh, K.; Harada, M. Identification of new prostate stem cell antigen-derived peptides immunogenic in HLA-A2(+) patients with hormone-refractory prostate cancer. Cancer Immunol. Immunother. 2004, 53, 479–489. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baaten, B.J.; Li, C.R.; Deiro, M.F.; Lin, M.M.; Linton, P.J.; Bradley, L.M. CD44 regulates survival and memory development in Th1 cells. Immunity 2010, 32, 104–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumann, J.; Stanko, K.; Schliesser, U.; Appelt, C.; Sawitzki, B. Differences in CD44 Surface Expression Levels and Function Discriminates IL-17 and IFN-γ Producing Helper T Cells. PLoS ONE 2015, 10, e0132479. [Google Scholar] [CrossRef] [Green Version]
- Ying, J.; Tsujii, M.; Kondo, J.; Hayashi, Y.; Kato, M.; Akasaka, T.; Inoue, T.; Shiraishi, E.; Hiyama, S.; Tsujii, Y.; et al. The effectiveness of an anti-human IL-6 receptor monoclonal antibody combined with chemotherapy to target colon cancer stem-like cells. Int. J. Oncol. 2015, 46, 1551–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peitzsch, C.; Nathansen, J.; Schniewind, S.I.; Schwarz, F.; Dubrovska, A. Cancer Stem Cells in Head and Neck Squamous Cell Carcinoma: Identification, Characterization and Clinical Implications. Cancers 2019, 11, 616. [Google Scholar] [CrossRef] [Green Version]
- Hunter, C.A.; Jones, S.A. Corrigendum: IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2017, 18, 1271. [Google Scholar] [CrossRef] [Green Version]
- Beard, R.E.; Zheng, Z.; Lagisetty, K.H.; Burns, W.R.; Tran, E.; Hewitt, S.M.; Abate-Daga, D.; Rosati, S.F.; Fine, H.A.; Ferrone, S.; et al. Multiple chimeric antigen receptors successfully target chondroitin sulfate proteoglycan 4 in several different cancer histologies and cancer stem cells. J. Immunother. Cancer 2014, 2, 25. [Google Scholar] [CrossRef] [Green Version]
- Morgan, R.A.; Johnson, L.A.; Davis, J.L.; Zheng, Z.; Woolard, K.D.; Reap, E.A.; Feldman, S.A.; Chinnasamy, N.; Kuan, C.T.; Song, H.; et al. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Hum Gene. Ther. 2012, 23, 1043–1053. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.E.; Starr, R.; Aguilar, B.; Shami, A.F.; Martinez, C.; D’Apuzzo, M.; Barish, M.E.; Forman, S.J.; Jensen, M.C. Stem-like tumor-initiating cells isolated from IL13Rα2 expressing gliomas are targeted and killed by IL13-zetakine-redirected T Cells. Clin. Cancer Res. 2012, 18, 2199–2209. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Wu, Y.; Ma, W.; Zhang, S.; Zhang, Y.Q. Adoptive T-cell therapy of prostate cancer targeting the cancer stem cell antigen EpCAM. BMC Immunol. 2015, 16, 1. [Google Scholar] [CrossRef] [Green Version]
- Ilyas, S.; Yang, J.C. Landscape of Tumor Antigens in T Cell Immunotherapy. J. Immunol. 2015, 195, 5117–5122. [Google Scholar] [CrossRef] [Green Version]
- Yamada, R.; Takahashi, A.; Torigoe, T.; Morita, R.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; Kubo, T.; Watarai, K.; Kondo, T.; et al. Preferential expression of cancer/testis genes in cancer stem-like cells: Proposal of a novel sub-category, cancer/testis/stem gene. Tissue Antigens 2013, 81, 428–434. [Google Scholar] [CrossRef]
- Wei, Z.; Zhou, C.; Zhang, Z.; Guan, M.; Zhang, C.; Liu, Z.; Liu, Q. The Landscape of Tumor Fusion Neoantigens: A Pan-Cancer Analysis. Science 2019, 21, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Knochelmann, H.M.; Smith, A.S.; Dwyer, C.J.; Wyatt, M.M.; Mehrotra, S.; Paulos, C.M. CAR T Cells in Solid Tumors: Blueprints for Building Effective Therapies. Front. Immunol. 2018, 9, 1740. [Google Scholar] [CrossRef] [Green Version]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Springuel, L.; Lonez, C.; Alexandre, B.; Van Cutsem, E.; Machiels, J.H.; Van Den Eynde, M.; Prenen, H.; Hendlisz, A.; Shaza, L.; Carrasco, J.; et al. Chimeric Antigen Receptor-T Cells for Targeting Solid Tumors: Current Challenges and Existing Strategies. BioDrugs 2019, 33, 515–537. [Google Scholar] [CrossRef] [Green Version]
- Dotti, G.; Savoldo, B.; Brenner, M. Fifteen years of gene therapy based on chimeric antigen receptors: “Are we nearly there yet?”. Hum. Gene. Ther. 2009, 20, 1229–1239. [Google Scholar] [CrossRef]
- Eshhar, Z.; Waks, T.; Gross, G.; Schindler, D.G. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc. Natl. Acad. Sci. USA 1993, 90, 720–724. [Google Scholar] [CrossRef] [Green Version]
- Sadelain, M.; Rivière, I.; Riddell, S. Therapeutic T cell engineering. Nature 2017, 545, 423–431. [Google Scholar] [CrossRef] [Green Version]
- Visus, C.; Wang, Y.; Lozano-Leon, A.; Ferris, R.L.; Silver, S.; Szczepanski, M.J.; Brand, R.E.; Ferrone, C.R.; Whiteside, T.L.; Ferrone, S.; et al. Targeting ALDH(bright) human carcinoma-initiating cells with ALDH1A1-specific CD8+ T cells. Clin. Cancer Res. 2011, 17, 6174–6184. [Google Scholar] [CrossRef] [Green Version]
- Visus, C.; Ito, D.; Amoscato, A.; Maciejewska-Franczak, M.; Abdelsalem, A.; Dhir, R.; Shin, D.M.; Donnenberg, V.S.; Whiteside, T.L.; DeLeo, A.B. Identification of human aldehyde dehydrogenase 1 family member A1 as a novel CD8+ T-cell-defined tumor antigen in squamous cell carcinoma of the head and neck. Cancer Res. 2007, 67, 10538–10545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, S.; Kochin, V.; Kanaseki, T.; Hongo, A.; Tokita, S.; Kikuchi, Y.; Takaya, A.; Hirohashi, Y.; Tsukahara, T.; Terui, T.; et al. The Antigen ASB4 on Cancer Stem Cells Serves as a Target for CTL Immunotherapy of Colorectal Cancer. Cancer Immunol. Res. 2018, 6, 358–369. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Zeng, C.; Fang, C.; Seeruttun, S.R.; Lv, L.; Wang, W. A new strategy using ALDHhigh-CD8+T cells to inhibit tumorigenesis. PLoS ONE 2014, 9, e103193. [Google Scholar] [CrossRef] [Green Version]
- Volonté, A.; Di Tomaso, T.; Spinelli, M.; Todaro, M.; Sanvito, F.; Albarello, L.; Bissolati, M.; Ghirardelli, L.; Orsenigo, E.; Ferrone, S.; et al. Cancer-Initiating Cells from Colorectal Cancer Patients Escape from T Cell-Mediated Immunosurveillance In Vitro through Membrane-Bound IL-4. J. Immunol. 2014, 192, 523–532. [Google Scholar] [CrossRef] [Green Version]
- Morita, R.; Hirohashi, Y.; Torigoe, T.; Ito-Inoda, S.; Takahashi, A.; Mariya, T.; Asanuma, H.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; et al. Olfactory Receptor Family 7 Subfamily C Member 1 Is a Novel Marker of Colon Cancer-Initiating Cells and Is a Potent Target of Immunotherapy. Clin. Cancer Res. 2016, 22, 3298–3309. [Google Scholar] [CrossRef] [Green Version]
- Verdegaal, E.M.; de Miranda, N.F.; Visser, M.; Harryvan, T.; van Buuren, M.M.; Andersen, R.S.; Hadrup, S.R.; van der Minne, C.E.; Schotte, R.; Spits, H.; et al. Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 2016, 536, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Prasad, S.; Gaedicke, S.; Hettich, M.; Firat, E.; Niedermann, G. Patient-derived glioblastoma stem cells are killed by CD133-specific CAR T cells but induce the T cell aging marker CD57. Oncotarget 2015, 6, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Chow, K.K.; Naik, S.; Kakarla, S.; Brawley, V.S.; Shaffer, D.R.; Yi, Z.; Rainusso, N.; Wu, M.F.; Liu, H.; Kew, Y.; et al. T cells redirected to EphA2 for the immunotherapy of glioblastoma. Mol. Ther. 2013, 21, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Yi, Z.; Prinzing, B.L.; Cao, F.; Gottschalk, S.; Krenciute, G. Optimizing EphA2-CAR T Cells for the Adoptive Immunotherapy of Glioma. Mol. Ther. Methods Clin. Dev. 2018, 9, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Kurtova, A.V.; Vera, J.; Lu, A.; Bear, A.; Foster, A.E. Elimination of Cancer Stem Cells with Genetically Engineered T Cells Expressing a Stage Specific Embryonic Antigen-1 (SSEA1) Chimeric Antigen Receptor. Mol. Ther. 2010, S181. [Google Scholar]
- Ghazi, A.; Rainusso, N.; Salsman, V.; Heslop, H.; Gottschalk, S.; Ahmed, N. Targeting Cancer Stem Cells in Osteosarcoma. Mol. Ther. 2010, S181. [Google Scholar]
- Seitz, C.M.; Schroeder, S.; Knopf, P.; Krahl, A.C.; Hau, J.; Schleicher, S.; Martella, M.; Quintanilla-Martinez, L.; Kneilling, M.; Pichler, B.; et al. GD2-targeted chimeric antigen receptor T cells prevent metastasis formation by elimination of breast cancer stem-like cells. Oncoimmunology 2020, 9, 1683345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrd, T.T.; Fousek, K.; Pignata, A.; Szot, C.; Samaha, H.; Seaman, S.; Dobrolecki, L.; Salsman, V.S.; Oo, H.Z.; Bielamowicz, K.; et al. TEM8/ANTXR1-Specific CAR T Cells as a Targeted Therapy for Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 489–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgenroth, A.; Cartellieri, M.; Schmitz, M.; Günes, S.; Weigle, B.; Bachmann, M.; Abken, H.; Rieber, E.P.; Temme, A. Targeting of tumor cells expressing the prostate stem cell antigen (PSCA) using genetically engineered T-cells. Prostate 2007, 67, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Ang, W.X.; Li, Z.; Chi, Z.; Du, S.H.; Chen, C.; Tay, J.C.; Toh, H.C.; Connolly, J.E.; Xu, X.H.; Wang, S. Intraperitoneal immunotherapy with T cells stably and transiently expressing anti-EpCAM CAR in xenograft models of peritoneal carcinomatosis. Oncotarget 2017, 8, 13545–13559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Feng, K.; Wang, Y.; Han, W. Targeting cancer stem cells by using chimeric antigen receptor-modified T cells: A potential and curable approach for cancer treatment. Protein Cell 2018, 9, 516–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sureban, S.M.; Berahovich, R.; Zhou, H.; Xu, S.; Wu, L.; Ding, K.; May, R.; Qu, D.; Bannerman-Menson, E.; Golubovskaya, V.; et al. DCLK1 Monoclonal Antibody-Based CAR-T Cells as a Novel Treatment Strategy against Human Colorectal Cancers. Cancers 2019, 12, 54. [Google Scholar] [CrossRef] [Green Version]
- Delgiorno, K.E.; Hall, J.C.; Takeuchi, K.K.; Pan, F.C.; Halbrook, C.J.; Washington, M.K.; Olive, K.P.; Spence, J.R.; Sipos, B.; Wright, C.V.; et al. Identification and manipulation of biliary metaplasia in pancreatic tumors. Gastroenterology 2014, 146, 233–244.e235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, J.M.; Alsina, J.; Rasheed, Z.A.; McAllister, F.M.; Fu, Y.Y.; Plentz, R.; Zhang, H.; Pasricha, P.J.; Bardeesy, N.; Matsui, W.; et al. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology 2014, 146, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi, Y.; Seno, H.; Fukuoka, A.; Ueo, T.; Yamaga, Y.; Maruno, T.; Nakanishi, N.; Kanda, K.; Komekado, H.; Kawada, M.; et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 2013, 45, 98–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaccari, P.; Cardinale, V.; Severi, C.; Pedica, F.; Carpino, G.; Gaudio, E.; Doglioni, C.; Petrone, M.C.; Alvaro, D.; Arcidiacono, P.G.; et al. Common features between neoplastic and preneoplastic lesions of the biliary tract and the pancreas. World J. Gastroenterol. 2019, 25, 4343–4359. [Google Scholar] [CrossRef] [PubMed]
- Hillerdal, V.; Ramachandran, M.; Leja, J.; Essand, M. Systemic treatment with CAR-engineered T cells against PSCA delays subcutaneous tumor growth and prolongs survival of mice. BMC Cancer 2014, 14, 30. [Google Scholar] [CrossRef] [Green Version]
- Priceman, S.J.; Gerdts, E.A.; Tilakawardane, D.; Kennewick, K.T.; Murad, J.P.; Park, A.K.; Jeang, B.; Yamaguchi, Y.; Yang, X.; Urak, R.; et al. Co-stimulatory signaling determines tumor antigen sensitivity and persistence of CAR T cells targeting PSCA+ metastatic prostate cancer. Oncoimmunology 2018, 7, e1380764. [Google Scholar] [CrossRef] [PubMed]
- Kloss, C.C.; Condomines, M.; Cartellieri, M.; Bachmann, M.; Sadelain, M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 2013, 31, 71–75. [Google Scholar] [CrossRef]
- Feldmann, A.; Arndt, C.; Bergmann, R.; Loff, S.; Cartellieri, M.; Bachmann, D.; Aliperta, R.; Hetzenecker, M.; Ludwig, F.; Albert, S.; et al. Retargeting of T lymphocytes to PSCA- or PSMA positive prostate cancer cells using the novel modular chimeric antigen receptor platform technology “UniCAR”. Oncotarget 2017, 8, 31368–31385. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, D.M.; Nasrallah, M.P.; Desai, A.; Melenhorst, J.J.; Mansfield, K.; Morrissette, J.J.D.; Martinez-Lage, M.; Brem, S.; Maloney, E.; Shen, A.; et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goff, S.L.; Morgan, R.A.; Yang, J.C.; Sherry, R.M.; Robbins, P.F.; Restifo, N.P.; Feldman, S.A.; Lu, Y.C.; Lu, L.; Zheng, Z.; et al. Pilot Trial of Adoptive Transfer of Chimeric Antigen Receptor-transduced T Cells Targeting EGFRvIII in Patients with Glioblastoma. J. Immunother. 2019, 42, 126–135. [Google Scholar] [CrossRef]
- Feng, K.C.; Guo, Y.L.; Liu, Y.; Dai, H.R.; Wang, Y.; Lv, H.Y.; Huang, J.H.; Yang, Q.M.; Han, W.D. Cocktail treatment with EGFR-specific and CD133-specific chimeric antigen receptor-modified T cells in a patient with advanced cholangiocarcinoma. J. Hematol. Oncol. 2017, 10, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.E.; Alizadeh, D.; Starr, R.; Weng, L.; Wagner, J.R.; Naranjo, A.; Ostberg, J.R.; Blanchard, M.S.; Kilpatrick, J.; Simpson, J.; et al. Regression of Glioblastoma after Chimeric Antigen Receptor T-Cell Therapy. N. Engl. J. Med. 2016, 375, 2561–2569. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, M.; Wu, Z.; Tong, C.; Dai, H.; Guo, Y.; Liu, Y.; Huang, J.; Lv, H.; Luo, C.; et al. CD133-directed CAR T cells for advanced metastasis malignancies: A phase I trial. Oncoimmunology 2018, 7, e1440169. [Google Scholar] [CrossRef] [Green Version]
- Pittari, G.; Filippini, P.; Gentilcore, G.; Grivel, J.C.; Rutella, S. Revving up Natural Killer Cells and Cytokine-Induced Killer Cells Against Hematological Malignancies. Front. Immunol. 2015, 6, 230. [Google Scholar] [CrossRef] [Green Version]
- Moretta, A.; Marcenaro, E.; Parolini, S.; Ferlazzo, G.; Moretta, L. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008, 15, 226–233. [Google Scholar] [CrossRef] [Green Version]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Morvan, M.G.; Lanier, L.L. NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 2016, 16, 7–19. [Google Scholar] [CrossRef]
- Wang, W.; Erbe, A.K.; Hank, J.A.; Morris, Z.S.; Sondel, P.M. NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Cancer Immunotherapy. Front. Immunol. 2015, 6, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezvani, K.; Rouce, R.; Liu, E.; Shpall, E. Engineering Natural Killer Cells for Cancer Immunotherapy. Mol. Ther. 2017, 25, 1769–1781. [Google Scholar] [CrossRef]
- Jewett, A.; Man, Y.G.; Tseng, H.C. Dual functions of natural killer cells in selection and differentiation of stem cells; role in regulation of inflammation and regeneration of tissues. J. Cancer 2013, 4, 12–24. [Google Scholar] [CrossRef]
- Levy, E.M.; Roberti, M.P.; Mordoh, J. Natural killer cells in human cancer: From biological functions to clinical applications. J. Biomed. Biotechnol. 2011, 2011, 676198. [Google Scholar] [CrossRef]
- Cheng, M.; Chen, Y.; Xiao, W.; Sun, R.; Tian, Z. NK cell-based immunotherapy for malignant diseases. Cell Mol. Immunol. 2013, 10, 230–252. [Google Scholar] [CrossRef] [PubMed]
- Granzin, M.; Wagner, J.; Köhl, U.; Cerwenka, A.; Huppert, V.; Ullrich, E. Shaping of Natural Killer Cell Antitumor Activity by. Front. Immunol. 2017, 8, 458. [Google Scholar] [CrossRef] [Green Version]
- Klingemann, H.; Boissel, L.; Toneguzzo, F. Natural Killer Cells for Immunotherapy-Advantages of the NK-92 Cell Line over Blood NK Cells. Front. Immunol. 2016, 7, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tam, Y.K.; Martinson, J.A.; Doligosa, K.; Klingemann, H.G. Ex vivo expansion of the highly cytotoxic human natural killer-92 cell-line under current good manufacturing practice conditions for clinical adoptive cellular immunotherapy. Cytotherapy 2003, 5, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Suck, G.; Odendahl, M.; Nowakowska, P.; Seidl, C.; Wels, W.S.; Klingemann, H.G.; Tonn, T. NK-92: An ‘off-the-shelf therapeutic’ for adoptive natural killer cell-based cancer immunotherapy. Cancer Immunol. Immunother. 2016, 65, 485–492. [Google Scholar] [CrossRef]
- Tallerico, R.; Garofalo, C.; Carbone, E. A New Biological Feature of Natural Killer Cells: The Recognition of Solid Tumor-Derived Cancer Stem Cells. Front. Immunol. 2016, 7, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossenbacher, S.K.; Canter, R.J.; Murphy, W.J. Natural killer cell immunotherapy to target stem-like tumor cells. J. Immunother Cancer 2016, 4, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Soto, A.; Gonzalez, S.; Smyth, M.J.; Galluzzi, L. Control of Metastasis by NK Cells. Cancer Cell 2017, 32, 135–154. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. Expression and function of immune ligand-receptor pairs in NK cells and cancer stem cells: Therapeutic implications. Cell Oncol. (Dordr) 2018, 41, 107–121. [Google Scholar] [CrossRef]
- Chiou, S.H.; Wang, M.L.; Chou, Y.T.; Chen, C.J.; Hong, C.F.; Hsieh, W.J.; Chang, H.T.; Chen, Y.S.; Lin, T.W.; Hsu, H.S.; et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010, 70, 10433–10444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, H.C.; Kanayama, K.; Kaur, K.; Park, S.H.; Park, S.; Kozlowska, A.; Sun, S.; McKenna, C.E.; Nishimura, I.; Jewett, A. Erratum: Bisphosphonate-induced differential modulation of immune cell function in gingiva and bone marrow in vivo: Role in osteoclast-mediated NK cell activation. Oncotarget 2015, 6, 41398. [Google Scholar] [CrossRef]
- Kozlowska, A.K.; Topchyan, P.; Kaur, K.; Tseng, H.C.; Teruel, A.; Hiraga, T.; Jewett, A. Differentiation by NK cells is a prerequisite for effective targeting of cancer stem cells/poorly differentiated tumors by chemopreventive and chemotherapeutic drugs. J. Cancer 2017, 8, 537–554. [Google Scholar] [CrossRef]
- Castriconi, R.; Daga, A.; Dondero, A.; Zona, G.; Poliani, P.L.; Melotti, A.; Griffero, F.; Marubbi, D.; Spaziante, R.; Bellora, F.; et al. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J. Immunol. 2009, 182, 3530–3539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ames, E.; Canter, R.J.; Grossenbacher, S.K.; Mac, S.; Chen, M.; Smith, R.C.; Hagino, T.; Perez-Cunningham, J.; Sckisel, G.D.; Urayama, S.; et al. NK Cells Preferentially Target Tumor Cells with a Cancer Stem Cell Phenotype. J. Immunol. 2015, 195, 4010–4019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, T.; Wang, G.; He, S.; Liu, Q.; Sun, J.; Wang, Y. Human cancer cells with stem cell-like phenotype exhibit enhanced sensitivity to the cytotoxicity of IL-2 and IL-15 activated natural killer cells. Cell Immunol. 2016, 300, 41–45. [Google Scholar] [CrossRef]
- Tseng, H.C.; Arasteh, A.; Paranjpe, A.; Teruel, A.; Yang, W.; Behel, A.; Alva, J.A.; Walter, G.; Head, C.; Ishikawa, T.O.; et al. Increased lysis of stem cells but not their differentiated cells by natural killer cells; de-differentiation or reprogramming activates NK cells. PLoS ONE 2010, 5, e11590. [Google Scholar] [CrossRef] [Green Version]
- Tallerico, R.; Todaro, M.; Di Franco, S.; Maccalli, C.; Garofalo, C.; Sottile, R.; Palmieri, C.; Tirinato, L.; Pangigadde, P.N.; La Rocca, R.; et al. Human NK cells selective targeting of colon cancer-initiating cells: A role for natural cytotoxicity receptors and MHC class I molecules. J. Immunol. 2013, 190, 2381–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietra, G.; Manzini, C.; Vitale, M.; Balsamo, M.; Ognio, E.; Boitano, M.; Queirolo, P.; Moretta, L.; Mingari, M.C. Natural killer cells kill human melanoma cells with characteristics of cancer stem cells. Int. Immunol. 2009, 21, 793–801. [Google Scholar] [CrossRef]
- Luna, J.I.; Grossenbacher, S.K.; Murphy, W.J.; Canter, R.J. Targeting Cancer Stem Cells with Natural Killer Cell Immunotherapy. Expert Opin. Biol. Ther. 2017, 17, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Konjević, G.M.; Vuletić, A.M.; Mirjačić Martinović, K.M.; Larsen, A.K.; Jurišić, V.B. The role of cytokines in the regulation of NK cells in the tumor environment. Cytokine 2019, 117, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Wiesner, S.; Xiao, J.; Ericson, K.; Chen, W.; Hall, W.A.; Low, W.C.; Ohlfest, J.R. Expression of MHC I and NK ligands on human CD133+ glioma cells: Possible targets of immunotherapy. J. Neurooncol. 2007, 83, 121–131. [Google Scholar] [CrossRef]
- Ferreira-Teixeira, M.; Paiva-Oliveira, D.; Parada, B.; Alves, V.; Sousa, V.; Chijioke, O.; Münz, C.; Reis, F.; Rodrigues-Santos, P.; Gomes, C. Natural killer cell-based adoptive immunotherapy eradicates and drives differentiation of chemoresistant bladder cancer stem-like cells. BMC Med. 2016, 14, 163. [Google Scholar] [CrossRef] [Green Version]
- Kaur, K.; Cook, J.; Park, S.H.; Topchyan, P.; Kozlowska, A.; Ohanian, N.; Fang, C.; Nishimura, I.; Jewett, A. Novel Strategy to Expand Super-Charged NK Cells with Significant Potential to Lyse and Differentiate Cancer Stem Cells: Differences in NK Expansion and Function between Healthy and Cancer Patients. Front. Immunol. 2017, 8, 297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jewett, A.; Kos, J.; Kaur, K.; Safaei, T.; Sutanto, C.; Chen, W.; Wong, P.; Namagerdi, A.K.; Fang, C.; Fong, Y.; et al. Natural Killer Cells: Diverse Functions in Tumor Immunity and Defects in Pre-neoplastic and Neoplastic Stages of Tumorigenesis. Mol. Ther. Oncolytics 2020, 16, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Tseng, H.C.; Bui, V.; Man, Y.G.; Cacalano, N.; Jewett, A. Induction of Split Anergy Conditions Natural Killer Cells to Promote Differentiation of Stem Cells through Cell-Cell Contact and Secreted Factors. Front. Immunol. 2014, 5, 269. [Google Scholar] [CrossRef] [Green Version]
- Deonarain, M.P.; Kousparou, C.A.; Epenetos, A.A. Antibodies targeting cancer stem cells: A new paradigm in immunotherapy? MAbs 2009, 1, 12–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmohl, J.U.; Felices, M.; Todhunter, D.; Taras, E.; Miller, J.S.; Vallera, D.A. Tetraspecific scFv construct provides NK cell mediated ADCC and self-sustaining stimuli via insertion of IL-15 as a cross-linker. Oncotarget 2016, 7, 73830–73844. [Google Scholar] [CrossRef] [PubMed]
- Dianat-Moghadam, H.; Rokni, M.; Marofi, F.; Panahi, Y.; Yousefi, M. Natural killer cell-based immunotherapy: From transplantation toward targeting cancer stem cells. J. Cell Physiol. 2018, 234, 259–273. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Hermanson, D.L.; Moriarity, B.S.; Kaufman, D.S. Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell Stem Cell 2018, 23, 181–192.e185. [Google Scholar] [CrossRef] [Green Version]
- Liu, E.; Tong, Y.; Dotti, G.; Shaim, H.; Savoldo, B.; Mukherjee, M.; Orange, J.; Wan, X.; Lu, X.; Reynolds, A.; et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 2018, 32, 520–531. [Google Scholar] [CrossRef]
- Müller, N.; Michen, S.; Tietze, S.; Töpfer, K.; Schulte, A.; Lamszus, K.; Schmitz, M.; Schackert, G.; Pastan, I.; Temme, A. Engineering NK Cells Modified With an EGFRvIII-specific Chimeric Antigen Receptor to Overexpress CXCR4 Improves Immunotherapy of CXCL12/SDF-1α-secreting Glioblastoma. J. Immunother. 2015, 38, 197–210. [Google Scholar] [CrossRef] [Green Version]
- Altvater, B.; Landmeier, S.; Pscherer, S.; Temme, J.; Schweer, K.; Kailayangiri, S.; Campana, D.; Juergens, H.; Pule, M.; Rossig, C. 2B4 (CD244) signaling by recombinant antigen-specific chimeric receptors costimulates natural killer cell activation to leukemia and neuroblastoma cells. Clin. Cancer Res. 2009, 15, 4857–4866. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Wang, G.; Huang, D.; Sui, M.; Xu, Y. Cancer Immunotherapy Based on Natural Killer Cells: Current Progress and New Opportunities. Front. Immunol. 2019, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Kailayangiri, S.; Altvater, B.; Spurny, C.; Jamitzky, S.; Schelhaas, S.; Jacobs, A.H.; Wiek, C.; Roellecke, K.; Hanenberg, H.; Hartmann, W.; et al. Targeting Ewing sarcoma with activated and GD2-specific chimeric antigen receptor-engineered human NK cells induces upregulation of immune-inhibitory HLA-G. Oncoimmunology 2017, 6, e1250050. [Google Scholar] [CrossRef] [Green Version]
- Golinelli, G.; Grisendi, G.; Prapa, M.; Bestagno, M.; Spano, C.; Rossignoli, F.; Bambi, F.; Sardi, I.; Cellini, M.; Horwitz, E.M.; et al. Targeting GD2-positive glioblastoma by chimeric antigen receptor empowered mesenchymal progenitors. Cancer Gene Ther. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidel, D.; Shibina, A.; Siebert, N.; Wels, W.S.; Reynolds, C.P.; Huebener, N.; Lode, H.N. Disialoganglioside-specific human natural killer cells are effective against drug-resistant neuroblastoma. Cancer Immunol. Immunother. 2015, 64, 621–634. [Google Scholar] [CrossRef]
- Burger, M.C.; Zhang, C.; Harter, P.N.; Romanski, A.; Strassheimer, F.; Senft, C.; Tonn, T.; Steinbach, J.P.; Wels, W.S. CAR-Engineered NK Cells for the Treatment of Glioblastoma: Turning Innate Effectors Into Precision Tools for Cancer Immunotherapy. Front. Immunol. 2019, 10, 2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haspels, H.N.; Rahman, M.A.; Joseph, J.V.; Gras Navarro, A.; Chekenya, M. Glioblastoma Stem-Like Cells Are More Susceptible Than Differentiated Cells to Natural Killer Cell Lysis Mediated Through Killer Immunoglobulin-Like Receptors-Human Leukocyte Antigen Ligand Mismatch and Activation Receptor-Ligand Interactions. Front. Immunol. 2018, 9, 1345. [Google Scholar] [CrossRef]
- Boissel, L.; Betancur-Boissel, M.; Lu, W.; Krause, D.S.; Van Etten, R.A.; Wels, W.S.; Klingemann, H. Retargeting NK-92 cells by means of CD19- and CD20-specific chimeric antigen receptors compares favorably with antibody-dependent cellular cytotoxicity. Oncoimmunology 2013, 2, e26527. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Burger, M.C.; Jennewein, L.; Genßler, S.; Schönfeld, K.; Zeiner, P.; Hattingen, E.; Harter, P.N.; Mittelbronn, M.; Tonn, T.; et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Klapdor, R.; Wang, S.; Hacker, U.; Büning, H.; Morgan, M.; Dörk, T.; Hillemanns, P.; Schambach, A. Improved Killing of Ovarian Cancer Stem Cells by Combining a Novel Chimeric Antigen Receptor-Based Immunotherapy and Chemotherapy. Hum. Gene Ther. 2017, 28, 886–896. [Google Scholar] [CrossRef]
- Burger, M.C.; Mildenberger, I.C.; Cieplik, H.C.; Zhang, C.; Jennewein, L.; Ihrig, K.; Wagner, M.; Mittelbronn, M.; Senft, C.; Tonn, T.; et al. The CAR2BRAIN study: A monocentric phase I trial with ErbB2-specific NK-92/5.28.z cells in recurrent glioblastoma. Neuro-Oncol. 2017, 19, iii51–iii52. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zheng, H.; Diao, Y. Natural Killer Cells and Current Applications of Chimeric Antigen Receptor-Modified NK-92 Cells in Tumor Immunotherapy. Int. J. Mol. Sci. 2019, 20, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.H.; Negrin, R.S. A novel population of expanded human CD3+CD56+ cells derived from T cells with potent in vivo antitumor activity in mice with severe combined immunodeficiency. J. Immunol. 1994, 153, 1687–1696. [Google Scholar] [PubMed]
- Baker, J.; Verneris, M.R.; Ito, M.; Shizuru, J.A.; Negrin, R.S. Expansion of cytolytic CD8(+) natural killer T cells with limited capacity for graft-versus-host disease induction due to interferon gamma production. Blood 2001, 97, 2923–2931. [Google Scholar] [CrossRef] [PubMed]
- Todorovic, M.; Mesiano, G.; Gammaitoni, L.; Leuci, V.; Giraudo, L.; Cammarata, C.; Jordaney, N.; Carnevale-Schianca, F.; Gallo, S.; Fagioli, F.; et al. Ex vivo allogeneic stimulation significantly improves expansion of cytokine-induced killer cells without increasing their alloreactivity across HLA barriers. J. Immunother. 2012, 35, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Mesiano, G.; Todorovic, M.; Gammaitoni, L.; Leuci, V.; Giraudo Diego, L.; Carnevale-Schianca, F.; Fagioli, F.; Piacibello, W.; Aglietta, M.; Sangiolo, D. Cytokine-induced killer (CIK) cells as feasible and effective adoptive immunotherapy for the treatment of solid tumors. Expert Opin. Biol. Ther. 2012, 12, 673–684. [Google Scholar] [CrossRef]
- Leuci, V.; Mesiano, G.; Gammaitoni, L.; Aglietta, M.; Sangiolo, D. Genetically Redirected T Lymphocytes for Adoptive Immunotherapy of Solid Tumors. Curr. Gene Ther. 2014, 14, 52–62. [Google Scholar] [CrossRef] [Green Version]
- Cappuzzello, E.; Sommaggio, R.; Zanovello, P.; Rosato, A. Cytokines for the induction of antitumor effectors: The paradigm of Cytokine-Induced Killer (CIK) cells. Cytokine Growth Factor Rev. 2017, 36, 99–105. [Google Scholar] [CrossRef]
- Schmidt-Wolf, I.G.; Lefterova, P.; Mehta, B.A.; Fernandez, L.P.; Huhn, D.; Blume, K.G.; Weissman, I.L.; Negrin, R.S. Phenotypic characterization and identification of effector cells involved in tumor cell recognition of cytokine-induced killer cells. Exp. Hematol. 1993, 21, 1673–1679. [Google Scholar]
- Palmerini, P.; Dalla Pietà, A.; Sommaggio, R.; Ventura, A.; Astori, G.; Chieregato, K.; Tisi, M.C.; Visco, C.; Perbellini, O.; Ruggeri, M.; et al. A serum-free protocol for the ex vivo expansion of Cytokine-Induced Killer cells using gas-permeable static culture flasks. Cytotherapy 2020, 22, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Introna, M.; Correnti, F. Innovative Clinical Perspectives for CIK Cells in Cancer Patients. Int. J. Mol. Sci. 2018, 19, 358. [Google Scholar] [CrossRef] [Green Version]
- Pende, D.; Rivera, P.; Marcenaro, S.; Chang, C.C.; Biassoni, R.; Conte, R.; Kubin, M.; Cosman, D.; Ferrone, S.; Moretta, L.; et al. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: Analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002, 62, 6178–6186. [Google Scholar]
- Franceschetti, M.; Pievani, A.; Borleri, G.; Vago, L.; Fleischhauer, K.; Golay, J.; Introna, M. Cytokine-induced killer cells are terminally differentiated activated CD8 cytotoxic T-EMRA lymphocytes. Exp. Hematol. 2009, 37, 616–628.e612. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, A.; Jamieson, A.M.; Liu, S.D.; Shastri, N.; Raulet, D.H. Ligands for the murine NKG2D receptor: Expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 2000, 1, 119–126. [Google Scholar] [CrossRef]
- Verneris, M.R.; Karami, M.; Baker, J.; Jayaswal, A.; Negrin, R.S. Role of NKG2D signaling in the cytotoxicity of activated and expanded CD8+ T cells. Blood 2004, 103, 3065–3072. [Google Scholar] [CrossRef] [Green Version]
- Schmeel, L.C.; Schmeel, F.C.; Coch, C.; Schmidt-Wolf, I.G. Cytokine-induced killer (CIK) cells in cancer immunotherapy: Report of the international registry on CIK cells (IRCC). J. Cancer Res. Clin. Oncol. 2015, 141, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Olioso, P.; Giancola, R.; Di Riti, M.; Contento, A.; Accorsi, P.; Iacone, A. Immunotherapy with cytokine induced killer cells in solid and hematopoietic tumours: A pilot clinical trial. Hematol. Oncol. 2009, 27, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Wolf, I.G.; Finke, S.; Trojaneck, B.; Denkena, A.; Lefterova, P.; Schwella, N.; Heuft, H.G.; Prange, G.; Korte, M.; Takeya, M.; et al. Phase I clinical study applying autologous immunological effector cells transfected with the interleukin-2 gene in patients with metastatic renal cancer, colorectal cancer and lymphoma. Br. J. Cancer 1999, 81, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Leemhuis, T.; Wells, S.; Scheffold, C.; Edinger, M.; Negrin, R.S. A phase I trial of autologous cytokine-induced killer cells for the treatment of relapsed Hodgkin disease and non-Hodgkin lymphoma. Biol. Blood Marrow Transplant. 2005, 11, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Wang, C.; Liu, L.; Du, C.; Cao, S.; Yu, J.; Wang, S.E.; Hao, X.; Ren, X.; Li, H. Autologous cytokine-induced killer cell immunotherapy in lung cancer: A phase II clinical study. Cancer Immunol. Immunother. 2012, 61, 2125–2133. [Google Scholar] [CrossRef]
- Wei, F.; Rong, X.X.; Xie, R.Y.; Jia, L.T.; Wang, H.Y.; Qin, Y.J.; Chen, L.; Shen, H.F.; Lin, X.L.; Yang, J.; et al. Cytokine-induced killer cells efficiently kill stem-like cancer cells of nasopharyngeal carcinoma via the NKG2D-ligands recognition. Oncotarget 2015, 6, 35023–35039. [Google Scholar] [CrossRef] [Green Version]
- Gammaitoni, L.; Giraudo, L.; Leuci, V.; Todorovic, M.; Mesiano, G.; Picciotto, F.; Pisacane, A.; Zaccagna, A.; Volpe, M.G.; Gallo, S.; et al. Effective Activity of Cytokine-Induced Killer Cells against Autologous Metastatic Melanoma Including Cells with Stemness Features. Clin. Cancer Res. 2013, 19, 4347–4358. [Google Scholar] [CrossRef] [Green Version]
- Sangiolo, D.; Mesiano, G.; Gammaitoni, L.; Leuci, V.; Todorovic, M.; Giraudo, L.; Cammarata, C.; Dell’aglio, C.; D’Ambrosio, L.; Pisacane, A.; et al. Cytokine-induced killer cells eradicate bone and soft-tissue sarcomas. Cancer Res. 2014, 74, 119–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rong, X.X.; Wei, F.; Lin, X.L.; Qin, Y.J.; Chen, L.; Wang, H.Y.; Shen, H.F.; Jia, L.T.; Xie, R.Y.; Lin, T.Y.; et al. Recognition and killing of cancer stem-like cell population in hepatocellular carcinoma cells by cytokine-induced killer cells via NKG2d-ligands recognition. Oncoimmunology 2016, 5, e1086060. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, Y.; Wang, Y.; Wu, D.; Lau, A.H.Y.; Zhao, P.; Zou, C.; Dai, Y.; Chan, F.L. Targeting prostate cancer stem-like cells by an immunotherapeutic platform based on immunogenic peptide-sensitized dendritic cells-cytokine-induced killer cells. Stem Cell Res. Ther. 2020, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Ma, W.; Lu, H.; Yuan, L.; An, L.; Wang, X.; Cheng, G.; Zuo, S. Modification of cytokine-induced killer cells with chimeric antigen receptors (CARs) enhances antitumor immunity to epidermal growth factor receptor (EGFR)-positive malignancies. Cancer Immunol. Immunother 2015, 64, 1517–1529. [Google Scholar] [CrossRef]
- Zuo, S.; Wen, Y.; Panha, H.; Dai, G.; Wang, L.; Ren, X.; Fu, K. Modification of cytokine-induced killer cells with folate receptor alpha (FRα)-specific chimeric antigen receptors enhances their antitumor immunity toward FRα-positive ovarian cancers. Mol. Immunol. 2017, 85, 293–304. [Google Scholar] [CrossRef]
- Merker, M.; Pfirrmann, V.; Oelsner, S.; Fulda, S.; Klingebiel, T.; Wels, W.S.; Bader, P.; Rettinger, E. Generation and characterization of ErbB2-CAR-engineered cytokine-induced killer cells for the treatment of high-risk soft tissue sarcoma in children. Oncotarget 2017, 8, 66137–66153. [Google Scholar] [CrossRef] [Green Version]
- Tettamanti, S.; Marin, V.; Pizzitola, I.; Magnani, C.F.; Giordano Attianese, G.M.; Cribioli, E.; Maltese, F.; Galimberti, S.; Lopez, A.F.; Biondi, A.; et al. Targeting of acute myeloid leukaemia by cytokine-induced killer cells redirected with a novel CD123-specific chimeric antigen receptor. Br. J. Haematol. 2013, 161, 389–401. [Google Scholar] [CrossRef]
- Pizzitola, I.; Anjos-Afonso, F.; Rouault-Pierre, K.; Lassailly, F.; Tettamanti, S.; Spinelli, O.; Biondi, A.; Biagi, E.; Bonnet, D. Chimeric antigen receptors against CD33/CD123 antigens efficiently target primary acute myeloid leukemia cells in vivo. Leukemia 2014, 28, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Merker, M.; Wagner, J.; Kreyenberg, H.; Heim, C.; Moser, L.M.; Wels, W.S.; Bonig, H.; Ivics, Z.; Ullrich, E.; Klingebiel, T.; et al. ERBB2-CAR-Engineered Cytokine-Induced Killer Cells Exhibit Both CAR-Mediated and Innate Immunity Against High-Risk Rhabdomyosarcoma. Front. Immunol. 2020, 11, 581468. [Google Scholar] [CrossRef]
- Magnani, C.F.; Gaipa, G.; Lussana, F.; Belotti, D.; Gritti, G.; Napolitano, S.; Matera, G.; Cabiati, B.; Buracchi, C.; Borleri, G.; et al. Sleeping Beauty-engineered CAR T cells achieve antileukemic activity without severe toxicities. J. Clin. Investig. 2020, 130, 6021–6033. [Google Scholar] [CrossRef]
- Guo, X.; Zheng, H.; Luo, W.; Zhang, Q.; Liu, J.; Yao, K. 5T4-specific chimeric antigen receptor modification promotes the immune efficacy of cytokine-induced killer cells against nasopharyngeal carcinoma stem cell-like cells. Sci. Rep. 2017, 7, 4859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolih, V.; Barutello, G.; Iussich, S.; De Maria, R.; Quaglino, E.; Buracco, P.; Cavallo, F.; Riccardo, F. CSPG4: A prototype oncoantigen for translational immunotherapy studies. J. Transl. Med. 2017, 15, 151. [Google Scholar] [CrossRef]
- Godfrey, D.I.; Uldrich, A.P.; McCluskey, J.; Rossjohn, J.; Moody, D.B. The burgeoning family of unconventional T cells. Nat. Immunol. 2015, 16, 1114–1123. [Google Scholar] [CrossRef]
- Salio, M.; Silk, J.D.; Jones, E.Y.; Cerundolo, V. Biology of CD1- and MR1-restricted T cells. Annu. Rev. Immunol. 2014, 32, 323–366. [Google Scholar] [CrossRef]
- Terabe, M.; Berzofsky, J.A. The immunoregulatory role of type I and type II NKT cells in cancer and other diseases. Cancer Immunol. Immunother. 2014, 63, 199–213. [Google Scholar] [CrossRef] [Green Version]
- Du, S.H.; Li, Z.; Chen, C.; Tan, W.K.; Chi, Z.; Kwang, T.W.; Xu, X.H.; Wang, S. Co-Expansion of Cytokine-Induced Killer Cells and Vγ9Vδ2 T Cells for CAR T-Cell Therapy. PLoS ONE 2016, 11, e0161820. [Google Scholar] [CrossRef] [PubMed]
- Heczey, A.; Liu, D.; Tian, G.; Courtney, A.N.; Wei, J.; Marinova, E.; Gao, X.; Guo, L.; Yvon, E.; Hicks, J.; et al. Invariant NKT cells with chimeric antigen receptor provide a novel platform for safe and effective cancer immunotherapy. Blood 2014, 124, 2824–2833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, B.; Wiesinger, M.; März, J.; Wistuba-Hamprecht, K.; Weide, B.; Schuler-Thurner, B.; Schuler, G.; Dörrie, J.; Uslu, U. The Generation of CAR-Transfected Natural Killer T Cells for the Immunotherapy of Melanoma. Int. J. Mol. Sci. 2018, 19, 2365. [Google Scholar] [CrossRef] [Green Version]
- Chien, Y.H.; Meyer, C.; Bonneville, M. γδ T cells: First line of defense and beyond. Annu. Rev. Immunol. 2014, 32, 121–155. [Google Scholar] [CrossRef]
- Vantourout, P.; Hayday, A. Six-of-the-best: Unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 2013, 13, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Groh, V.; Rhinehart, R.; Secrist, H.; Bauer, S.; Grabstein, K.H.; Spies, T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc. Natl. Acad. Sci. USA 1999, 96, 6879–6884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangan, B.A.; Dunne, M.R.; O’Reilly, V.P.; Dunne, P.J.; Exley, M.A.; O’Shea, D.; Scotet, E.; Hogan, A.E.; Doherty, D.G. Cutting edge: CD1d restriction and Th1/Th2/Th17 cytokine secretion by human Vδ3 T cells. J. Immunol. 2013, 191, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, F.; Tanaka, Y.; Yamashita, S.; Minato, N. Essential requirement of antigen presentation by monocyte lineage cells for the activation of primary human gamma delta T cells by aminobisphosphonate antigen. J. Immunol. 2001, 166, 5508–5514. [Google Scholar] [CrossRef]
- Sebestyen, Z.; Prinz, I.; Déchanet-Merville, J.; Silva-Santos, B.; Kuball, J. Translating gammadelta (γδ) T cells and their receptors into cancer cell therapies. Nat. Rev. Drug Discov. 2020, 19, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Todaro, M.; D’Asaro, M.; Caccamo, N.; Iovino, F.; Francipane, M.G.; Meraviglia, S.; Orlando, V.; La Mendola, C.; Gulotta, G.; Salerno, A.; et al. Efficient killing of human colon cancer stem cells by gammadelta T lymphocytes. J. Immunol. 2009, 182, 7287–7296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, D.; Wang, F.; Chen, Y.; Wang, C.; Liu, S.; Lu, B.; Ge, X.; Guo, L. Human ovarian cancer stem-like cells can be efficiently killed by γδ T lymphocytes. Cancer Immunol. Immunother. 2012, 61, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Nishio, N.; Fujita, M.; Tanaka, Y.; Maki, H.; Zhang, R.; Hirosawa, T.; Demachi-Okamura, A.; Uemura, Y.; Taguchi, O.; Takahashi, Y.; et al. Zoledronate sensitizes neuroblastoma-derived tumor-initiating cells to cytolysis mediated by human γδ T cells. J. Immunother. 2012, 35, 598–606. [Google Scholar] [CrossRef]
- Miyashita, M.; Tomogane, M.; Nakamura, Y.; Shimizu, T.; Fujihara, A.; Ukimura, O.; Ashihara, E. Sphere-derived Prostate Cancer Stem Cells Are Resistant to γδ T Cell Cytotoxicity. Anticancer Res. 2020, 40, 5481–5487. [Google Scholar] [CrossRef] [PubMed]
- Dutta, I.; Dieters-Castator, D.; Papatzimas, J.W.; Medina, A.; Schueler, J.; Derksen, D.J.; Lajoie, G.; Postovit, L.M.; Siegers, G.M. ADAM protease inhibition overcomes resistance of breast cancer stem-like cells to γδ T cell immunotherapy. Cancer Lett. 2021, 496, 156–168. [Google Scholar] [CrossRef]
- Nicol, A.J.; Tokuyama, H.; Mattarollo, S.R.; Hagi, T.; Suzuki, K.; Yokokawa, K.; Nieda, M. Clinical evaluation of autologous gamma delta T cell-based immunotherapy for metastatic solid tumours. Br. J. Cancer 2011, 105, 778–786. [Google Scholar] [CrossRef] [Green Version]
- Nada, M.H.; Wang, H.; Workalemahu, G.; Tanaka, Y.; Morita, C.T. Enhancing adoptive cancer immunotherapy with Vγ2Vδ2 T cells through pulse zoledronate stimulation. J. Immunother. Cancer 2017, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, T.; Matsushita, H.; Hoshikawa, M.; Hasegawa, K.; Kokudo, N.; Kakimi, K. Adjuvant combination therapy with gemcitabine and autologous γδ T-cell transfer in patients with curatively resected pancreatic cancer. Cytotherapy 2017, 19, 473–485. [Google Scholar] [CrossRef]
- Pressey, J.G.; Adams, J.; Harkins, L.; Kelly, D.; You, Z.; Lamb, L.S. In vivo expansion and activation of γδ T cells as immunotherapy for refractory neuroblastoma: A phase 1 study. Medicine (Baltimore) 2016, 95, e4909. [Google Scholar] [CrossRef]
- Chen, H.C.; Joalland, N.; Bridgeman, J.S.; Alchami, F.S.; Jarry, U.; Khan, M.W.A.; Piggott, L.; Shanneik, Y.; Li, J.; Herold, M.J.; et al. Synergistic targeting of breast cancer stem-like cells by human γδ T cells and CD8. Immunol. Cell Biol. 2017, 95, 620–629. [Google Scholar] [CrossRef] [Green Version]
- Lawson, D.A.; Kessenbrock, K.; Davis, R.T.; Pervolarakis, N.; Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 2018, 20, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Clara, J.A.; Monge, C.; Yang, Y.; Takebe, N. Targeting signalling pathways and the immune microenvironment of cancer stem cells-a clinical update. Nat. Rev. Clin. Oncol. 2020, 17, 204–232. [Google Scholar] [CrossRef] [PubMed]
- Adeegbe, D.O.; Liu, Y.; Lizotte, P.H.; Kamihara, Y.; Aref, A.R.; Almonte, C.; Dries, R.; Li, Y.; Liu, S.; Wang, X.; et al. Synergistic Immunostimulatory Effects and Therapeutic Benefit of Combined Histone Deacetylase and Bromodomain Inhibition in Non-Small Cell Lung Cancer. Cancer Discov. 2017, 7, 852–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gammaitoni, L.; Leuci, V.; Mesiano, G.; Giraudo, L.; Todorovic, M.; Carnevale-Schianca, F.; Aglietta, M.; Sangiolo, D. Immunotherapy of cancer stem cells in solid tumors: Initial findings and future prospective. Expert Opin. Biol. Ther. 2014, 14, 1259–1270. [Google Scholar] [CrossRef]
- Hombach, A.A.; Rappl, G.; Abken, H. Arming cytokine-induced killer cells with chimeric antigen receptors: CD28 outperforms combined CD28-OX40 “super-stimulation”. Mol. Ther. 2013, 21, 2268–2277. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.K.; Hamilton, C.A.; Cheung, M.K.; Karimi, M.; Baker, J.; Gall, J.M.; Schulz, S.; Thorne, S.H.; Teng, N.N.; Contag, C.H.; et al. Enhanced killing of primary ovarian cancer by retargeting autologous cytokine-induced killer cells with bispecific antibodies: A preclinical study. Clin. Cancer Res. 2006, 12, 1859–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Biological Agent | Strategy | Combination | Disease Target | NCT Identifier | Status | Phase |
|---|---|---|---|---|---|---|
| NK-cell based therapy | HER2-CAR.NK-92 (CAR2BRAIN) | / | Recurrent Glioblastoma | NCT03383978 | Recruiting | 1 |
| NKT-cell based therapy | GD2-CAR.NKT (GINAKIT) | Cyclophosphamide Fludarabine | Neuroblastoma | NCT03294954 | Recruiting | 1 |
| T-cell based therapy | IL13Rα2-CAR.T | / | Refractory Malignant Glioma | NCT02208362 | Recruiting | 1 |
| CD133-CAR.T | / | Liver Cancer Pancreatic Cancer Colorectal Cancer Brain Tumors Ovarian Cancer Breast Cancer | NCT02541370 | Completed | 1–2 | |
| EGFRvIII-CAR.T | Aldesleukin Cyclophosphamide Fludarabine | Malignant Glioma Glioblastoma Gliosarcoma | NCT01454596 | Completed | 1–2 | |
| EGFRvIII-CAR.T | / | Recurrent Glioma | NCT02209376 | Terminated | 1 | |
| EGFR-CAR.T plus CD133-CAR.T | / | Cholangiocarcinoma | / | Case Report | / | |
| MUC1-CAR.T PD-1 KO | / | Advanced Esophageal Cancer | NCT03706326 | Recruiting | 1–2 | |
| EGFR/IL-12-CAR.T | / | Metastatic Colorectal Cancer | NCT03542799 | Not Yet Recruiting | 1 | |
| MESO-CAR.T | / | Refractory Relapsed Ovarian Cancer | NCT03916679 | Recruiting | 1–2 | |
| MESO-19-CAR.T | / | Metastatic Pancreatic Cancer | NCT02465983 | Completed | 1 | |
| EpCAM-CAR.T | / | Recurrent Breast Cancer | NCT02915445 | Recruiting | 1 | |
| LeY-CAR.T | / | Advanced Cancer | NCT03851146 | Recruiting | 1 | |
| MOV19.BBz-CAR.T | / | Recurrent High-grade Serous Ovarian Cancer | NCT03585764 | Recruiting | 1 | |
| PSCA-CAR.T | Cyclophosphamide Fludarabine Fludarabine Phosfate | Castration-Resistant Prostate Carcinoma Metastatic Prostate Carcinoma Stage IV Prostate Cancer | NCT03873805 | Recruiting | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donini, C.; Rotolo, R.; Proment, A.; Aglietta, M.; Sangiolo, D.; Leuci, V. Cellular Immunotherapy Targeting Cancer Stem Cells: Preclinical Evidence and Clinical Perspective. Cells 2021, 10, 543. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10030543
Donini C, Rotolo R, Proment A, Aglietta M, Sangiolo D, Leuci V. Cellular Immunotherapy Targeting Cancer Stem Cells: Preclinical Evidence and Clinical Perspective. Cells. 2021; 10(3):543. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10030543
Chicago/Turabian StyleDonini, Chiara, Ramona Rotolo, Alessia Proment, Massimo Aglietta, Dario Sangiolo, and Valeria Leuci. 2021. "Cellular Immunotherapy Targeting Cancer Stem Cells: Preclinical Evidence and Clinical Perspective" Cells 10, no. 3: 543. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10030543