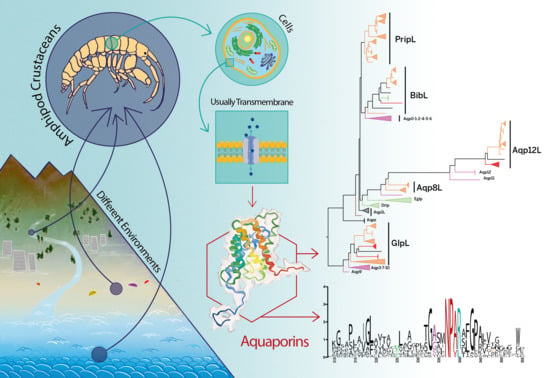
First Glimpse at the Diverse Aquaporins of Amphipod Crustaceans
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Phylogeny of the Translated Sequences
3.2. In Silico 3D Structures and Conserved Regions in Gammaroid AQPs
3.2.1. NPA Motifs
3.2.2. Ar/R Region
3.3. Use of AQPs for Phylogenetic Inferences: A Case Study of Gammaroid Amphipods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laloux, T.; Junqueira, B.; Maistriaux, L.C.; Ahmed, J.; Jurkiewicz, A.; Chaumont, F. Plant and Mammal Aquaporins: Same but Different. Int. J. Mol. Sci. 2018, 19, 521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michenkova, M.; Taki, S.; Blosser, M.C.; Hwang, H.J.; Kowatz, T.; Moss, F.J.; Occhipinti, R.; Qin, X.; Sen, S.; Shinn, E.; et al. Carbon dioxide transport across membranes. Interface Focus 2021, 11, 20200090. [Google Scholar] [CrossRef]
- Shapiguzov, A.Y. Aquaporins: Structure, Systematics, and Regulatory Features. Russ. J. Plant Physiol. 2004, 51, 127–137. [Google Scholar] [CrossRef]
- Fu, D.; Libson, A.; Larry, J.W.M.; Weitzman, C.; Nollert, P.; Krucinski, J.; Robert, M.S. Structure of a Glycerol-Conducting Channel and the Basis for Its Selectivity. Science 2000, 290, 481–486. [Google Scholar] [CrossRef] [Green Version]
- Hub, J.S.; de Groot, B.L. Mechanism of selectivity in aquaporins and aquaglyceroporins. Proc. Natl. Acad. Sci. USA 2008, 105, 1198. [Google Scholar] [CrossRef] [Green Version]
- Murata, K.; Mitsuoka, K.; Hirai, T.; Walz, T.; Agre, P.; Heymann, J.B.; Engel, A.; Fujiyoshi, Y. Structural determinants of water permeation through aquaporin-1. Nature 2000, 407, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Sui, H.; Han, B.-G.; Lee, J.K.; Walian, P.; Jap, B.K. Structural basis of water-specific transport through the AQP1 water channel. Nature 2001, 414, 872–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishibashi, K. Aquaporin subfamily with unusual NPA boxes. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 989–993. [Google Scholar] [CrossRef] [Green Version]
- Abascal, F.; Irisarri, I.; Zardoya, R. Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 1468–1481. [Google Scholar] [CrossRef] [Green Version]
- Soto, G.; Alleva, K.; Amodeo, G.; Muschietti, J.; Ayub, N.D. New insight into the evolution of aquaporins from flowering plants and vertebrates: Orthologous identification and functional transfer is possible. Gene 2012, 503, 165–176. [Google Scholar] [CrossRef]
- Ishibashi, K.; Tanaka, Y.; Morishita, Y. Chapter One—Perspectives on the evolution of aquaporin superfamily. Vitam. Horm. 2020, 112, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Benoit, J.B.; Hansen, I.A.; Szuter, E.M.; Drake, L.L.; Burnett, D.L.; Attardo, G.M. Emerging roles of aquaporins in relation to the physiology of blood-feeding arthropods. J. Comp. Physiol. B 2014, 184, 811–825. [Google Scholar] [CrossRef]
- Drake, L.L.; Boudko, D.Y.; Marinotti, O.; Carpenter, V.K.; Dawe, A.L.; Hansen, I.A. The Aquaporin Gene Family of the Yellow Fever Mosquito, Aedes aegypti. PLoS ONE 2011, 5, e15578. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Zhou, Y.; Jia, H.; Li, L.; Qian, J.; Han, F.; Yin, H.; Cui, Y. Transcriptomics-Based Identification of Aquaporin Diversity in the House Dust Mite Dermatophagoides farinae (Acariformes: Pyroglyphidae). J. Insect Sci. 2018, 18, 11. [Google Scholar] [CrossRef]
- Lind, U.; Järvå, M.; Rosenblad, M.A.; Pingitore, P.; Karlsson, E.; Wrange, A.-L.; Kamdal, E.; Sundell, K.; André, C.; Jonsson, P.R.; et al. Analysis of aquaporins from the euryhaline barnacle Balanus improvisus reveals differential expression in response to changes in salinity. PLoS ONE 2017, 12, e0181192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stavang, J.A.; Chauvigné, F.; Kongshaug, H.; Cerdà, J.; Nilsen, F.; Finn, R.N. Phylogenomic and functional analyses of salmon lice aquaporins uncover the molecular diversity of the superfamily in Arthropoda. BMC Genom. 2015, 16, 618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.N.; Cerdà, J. Evolution and Functional Diversity of Aquaporins. Biol. Bull. 2015, 229, 6–23. [Google Scholar] [CrossRef]
- Kaufmann, N.; Mathai, J.C.; Hill, W.G.; Dow, J.A.T.; Zeidel, M.L.; Brodsky, J.L. Developmental expression and biophysical characterization of a Drosophila melanogaster aquaporin. Am. J. Physiol.-Cell Physiol. 2005, 289, C397–C407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikawada, T.; Saito, A.; Kanamori, Y.; Fujita, M.; Śnigórska, K.; Watanabe, M.; Okuda, T. Dehydration-inducible changes in expression of two aquaporins in the sleeping chironomid, Polypedilum vanderplanki. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 514–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanochko, G.M.; Yool, A.J. Regulated Cationic Channel Function in Xenopus Oocytes Expressing Drosophila Big Brain. J. Neurosci. 2002, 22, 2530. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.N.; Chauvigné, F.; Stavang, J.A.; Belles, X.; Cerdà, J. Insect glycerol transporters evolved by functional co-option and gene replacement. Nat. Commun. 2015, 6, 7814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, J.; Hu, X.L.; Ip, J.C.H.; Ma, K.Y.; Tang, Y.; Wang, Y.; Qin, J.; Qiu, J.-W.; Chan, T.F.; Chu, K.H. Multi-omic approach provides insights into osmoregulation and osmoconformation of the crab Scylla paramamosain. Sci. Rep. 2020, 10, 21771. [Google Scholar] [CrossRef]
- Rahi, M.L.; Moshtaghi, A.; Mather, P.B.; Hurwood, D.A. Osmoregulation in decapod crustaceans: Physiological and genomic perspectives. Hydrobiologia 2018, 825, 177–188. [Google Scholar] [CrossRef]
- Horton, T.; Lowry, J.; De Broyer, C.; Bellan-Santini, D.; Coleman, C.O.; Corbari, L.; Costello, M.J.; Daneliya, M.; Dauvin, J.-C.; Fišer, C.; et al. World Amphipoda Database. 2021. Available online: http://www.marinespecies.org/amphipoda (accessed on 10 October 2021). [CrossRef]
- Väinölä, R.; Witt, J.D.S.; Grabowski, M.; Bradbury, J.H.; Jazdzewski, K.; Sket, B. Global diversity of amphipods (Amphipoda; Crustacea) in freshwater. Hydrobiologia 2008, 595, 241–255. [Google Scholar] [CrossRef]
- MacNeil, C.; Dick, J.T.A.; Platvoet, D.; Briffa, M. Direct and indirect effects of species displacements: An invading freshwater amphipod can disrupt leaf-litter processing and shredder efficiency. J. North Am. Benthol. Soc. 2011, 30, 38–48. [Google Scholar] [CrossRef]
- MacNeil, C.; Dick, J.T.A.; Elwood, R.W. The dynamics of predation on Gammarus spp. (Crustacea: Amphipoda). Biol. Rev. 1999, 74, 375–395. [Google Scholar] [CrossRef]
- Mamos, T.; Wattier, R.; Burzyński, A.; Grabowski, M. The legacy of a vanished sea: A high level of diversification within a European freshwater amphipod species complex driven by 15 My of Paratethys regression. Mol. Ecol. 2016, 25, 795–810. [Google Scholar] [CrossRef] [PubMed]
- Wildish, D.J. Evolutionary ecology of reproduction in gammaridean Amphipoda. Int. J. Invertebr. Reprod. 1982, 5, 1–19. [Google Scholar] [CrossRef]
- Grabowski, M.; Mamos, T.; Bącela-Spychalska, K.; Rewicz, T.; Wattier, R.A. Neogene paleogeography provides context for understanding the origin and spatial distribution of cryptic diversity in a widespread Balkan freshwater amphipod. PeerJ 2017, 5, e3016. [Google Scholar] [CrossRef] [Green Version]
- Wattier, R.; Mamos, T.; Copilaş-Ciocianu, D.; Jelić, M.; Ollivier, A.; Chaumot, A.; Danger, M.; Felten, V.; Piscart, C.; Žganec, K.; et al. Continental-scale patterns of hyper-cryptic diversity within the freshwater model taxon Gammarus fossarum (Crustacea, Amphipoda). Sci. Rep. 2020, 10, 16536. [Google Scholar] [CrossRef] [PubMed]
- Desiderato, A.; Costa, F.O.; Serejo, C.S.; Abbiati, M.; Queiroga, H.; Vieira, P.E. Macaronesian islands as promoters of diversification in amphipods: The remarkable case of the family Hyalidae (Crustacea, Amphipoda). Zool. Scr. 2019, 48, 359–375. [Google Scholar] [CrossRef]
- Grabowski, M.; Wysocka, A.; Mamos, T. Molecular species delimitation methods provide new insight into taxonomy of the endemic gammarid species flock from the ancient Lake Ohrid. Zool. J. Linn. Soc. 2017, 181, 272–285. [Google Scholar] [CrossRef]
- Gesteira, J.L.G.; Dauvin, J.C. Amphipods are Good Bioindicators of the Impact of Oil Spills on Soft-Bottom Macrobenthic Communities. Mar. Pollut. Bull. 2000, 40, 1017–1027. [Google Scholar] [CrossRef]
- Kunz, P.Y.; Kienle, C.; Gerhardt, A. Gammarus spp. in Aquatic Ecotoxicology and Water Quality Assessment: Toward Integrated Multilevel Tests. In Reviews of Environmental Contamination and Toxicology Volume 205; Whitacre, D.M., Ed.; Springer: New York, NY, USA, 2010; pp. 1–76. [Google Scholar] [CrossRef]
- Moore, P.G.; Rainbow, P.S.; Hayes, E. The beach-hopper Orchestia gammarellus (Crustacea: Amphipoda) as a biomonitor for copper and zinc: North Sea trials. Sci. Total. Environ. 1991, 106, 221–238. [Google Scholar] [CrossRef]
- Feckler, A.; Thielsch, A.; Schwenk, K.; Schulz, R.; Bundschuh, M. Differences in the sensitivity among cryptic lineages of the Gammarus fossarum complex. Sci. Total. Environ. 2012, 439, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Soucek, D.J.; Dickinson, A.; Major, K.M.; McEwen, A.R. Effect of test duration and feeding on relative sensitivity of genetically distinct clades of Hyalella azteca. Ecotoxicology 2013, 22, 1359–1366. [Google Scholar] [CrossRef]
- Conlan, K.E.; Desiderato, A.; Beermann, J. Jassa (Crustacea: Amphipoda): A new morphological and molecular assessment of the genus. Zootaxa 2021, 4939, 1–191. [Google Scholar] [CrossRef]
- Marchini, A.; Cardeccia, A. Alien amphipods in a sea of troubles: Cryptogenic species, unresolved taxonomy and overlooked introductions. Mar. Biol. 2017, 164, 69. [Google Scholar] [CrossRef]
- Cabezas, M.P.; Xavier, R.; Branco, M.; Santos, A.M.; Guerra-García, J.M. Invasion history of Caprella scaura Templeton, 1836 (Amphipoda: Caprellidae) in the Iberian Peninsula: Multiple introductions revealed by mitochondrial sequence data. Biol. Invasions 2014, 16, 2221–2245. [Google Scholar] [CrossRef]
- Rewicz, T.; Wattier, R.; Grabowski, M.; Rigaud, T.; Bacela-Spychalska, K. Out of the Black Sea: Phylogeography of the invasive killer shrimp Dikerogammarus villosus across Europe. PLoS ONE 2015, 10, e0118121. [Google Scholar] [CrossRef] [Green Version]
- Catalán-García, M.; Chauvigné, F.; Stavang, J.A.; Nilsen, F.; Cerdà, J.; Finn, R.N. Lineage-level divergence of copepod glycerol transporters and the emergence of isoform-specific trafficking regulation. Commun. Biol. 2021, 4, 643. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geneious 11.1. Available online: https://www.geneious.com (accessed on 1 September 2021).
- Mamos, T.; Grabowski, M.; Rewicz, T.; Bojko, J.; Strapagiel, D.; Burzyński, A. Mitochondrial Genomes, Phylogenetic Associations, and SNP Recovery for the Key Invasive Ponto-Caspian Amphipods in Europe. Int. J. Mol. Sci. 2021, 22, 300. [Google Scholar] [CrossRef]
- Pomianowski, K.; Burzyński, A.; Kulczykowska, E. A de novo Transcriptome Assembly of the European Flounder (Platichthys flesus): The Preselection of Transcripts Encoding Active Forms of Enzymes. Front. Mar. Sci. 2021, 8, 97. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anisimova, M.; Gascuel, O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Copilaş-Ciocianu, D.; Sidorov, D.; Gontcharov, A. Adrift across tectonic plates: Molecular phylogenetics supports the ancient Laurasian origin of old limnic crangonyctid amphipods. Org. Divers. Evol. 2019, 19, 191–207. [Google Scholar] [CrossRef]
- Copilaş-Ciocianu, D.; Zimţa, A.-A.; Grabowski, M.; Petrusek, A. Survival in northern microrefugia in an endemic Carpathian gammarid (Crustacea: Amphipoda). Zool. Scr. 2018, 47, 357–372. [Google Scholar] [CrossRef]
- Mamos, T.; Jażdżewski, K.; Čiamporová-Zaťovičová, Z.; Čiampor, F.J.; Grabowski, M. Fuzzy species borders of glacial survivalists in the Carpathian biodiversity hotspot revealed using a multimarker approach. Sci. Rep. 2021, 11, 21629. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, C.R.; Islam, S.A.; Sternberg, M.J.E. EzMol: A Web Server Wizard for the Rapid Visualization and Image Production of Protein and Nucleic Acid Structures. J. Mol. Biol. 2018, 430, 2244–2248. [Google Scholar] [CrossRef]
- Ittisoponpisan, S.; Islam, S.A.; Khanna, T.; Alhuzimi, E.; David, A.; Sternberg, M.J.E. Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated? J. Mol. Biol. 2019, 431, 2197–2212. [Google Scholar] [CrossRef]
- Verkman, A.S.; Yang, B.; Song, Y.; Manley, G.T.; Ma, T. Role of water channels in fluid transport studied by phenotype analysis of aquaporin knockout mice. Exp. Physiol. 2000, 85, 233s–241s. [Google Scholar] [CrossRef]
- WebLogo. Available online: https://weblogo.berkeley.edu/ (accessed on 1 October 2021).
- Ishibashi, K.; Morishita, Y.; Tanaka, Y. The Evolutionary Aspects of Aquaporin Family. In Aquaporins; Yang, B., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 2017; pp. 35–50. [Google Scholar] [CrossRef]
- Betts, M.J.; Russell, R.B. Amino Acid Properties and Consequences of Substitutions. Bioinform. Genet. 2003, 317, 289–316. [Google Scholar] [CrossRef]
- Danielson, J.Å.; Johanson, U. Unexpected complexity of the Aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.-X.; Pan, D.-D.; Xu, J.; Liu, Y.; Wang, G.-R.; Du, Y.-Z. Identification and Functional Analysis of the First Aquaporin from Striped Stem Borer, Chilo suppressalis. Front. Physiol. 2018, 9, 57. [Google Scholar] [CrossRef]
- Beitz, E.; Wu, B.; Holm, L.M.; Schultz, J.E.; Zeuthen, T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc. Natl. Acad. Sci. USA 2006, 103, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitchen, P.; Conner, A.C. Control of the Aquaporin-4 Channel Water Permeability by Structural Dynamics of Aromatic/Arginine Selectivity Filter Residues. Biochemistry 2015, 54, 6753–6755. [Google Scholar] [CrossRef]
- Rao, Y.; Jan, L.Y.; Jan, Y.N. Similarity of the product of the Drosophila neurogenic gene big brain to transmembrane channel proteins. Nature 1990, 345, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Doherty, D.; Jan, L.Y.; Jan, Y.N. The Drosophila neurogenic gene big brain, which encodes a membrane-associated protein, acts cell autonomously and can act synergistically with Notch and Delta. Development 1997, 124, 3881–3893. [Google Scholar] [CrossRef] [PubMed]
- Herraiz, A.; Chauvigné, F.; Cerdà, J.; Bellés, X.; Piulachs, M.-D. Identification and functional characterization of an ovarian aquaporin from the cockroach Blattella germanica L. (Dictyoptera, Blattellidae). J. Exp. Biol. 2011, 214, 3630–3638. [Google Scholar] [CrossRef] [Green Version]
- Kitchen, P.; Salman, M.M.; Pickel, S.U.; Jennings, J.; Törnroth-Horsefield, S.; Conner, M.T.; Bill, R.M.; Conner, A.C. Water channel pore size determines exclusion properties but not solute selectivity. Sci. Rep. 2019, 9, 20369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumenko, S.A.; Logacheva, M.D.; Popova, N.V.; Klepikova, A.V.; Penin, A.A.; Bazykin, G.A.; Etingova, A.E.; Mugue, N.S.; Kondrashov, A.S.; Yampolsky, L.Y. Transcriptome-based phylogeny of endemic Lake Baikal amphipod species flock: Fast speciation accompanied by frequent episodes of positive selection. Mol. Ecol. 2017, 26, 536–553. [Google Scholar] [CrossRef]
- Copilaş-Ciocianu, D.; Borko, Š.; Fišer, C. The late blooming amphipods: Global change promoted post-Jurassic ecological radiation despite Palaeozoic origin. Mol. Phylogenetics Evol. 2020, 143, 106664. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desiderato, A.; Mamos, T.; Rewicz, T.; Burzynski, A.; Mucciolo, S. First Glimpse at the Diverse Aquaporins of Amphipod Crustaceans. Cells 2021, 10, 3417. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10123417
Desiderato A, Mamos T, Rewicz T, Burzynski A, Mucciolo S. First Glimpse at the Diverse Aquaporins of Amphipod Crustaceans. Cells. 2021; 10(12):3417. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10123417
Chicago/Turabian StyleDesiderato, Andrea, Tomasz Mamos, Tomasz Rewicz, Artur Burzynski, and Serena Mucciolo. 2021. "First Glimpse at the Diverse Aquaporins of Amphipod Crustaceans" Cells 10, no. 12: 3417. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10123417