Molecular Mechanisms in Pentanucleotide Repeat Diseases
Abstract
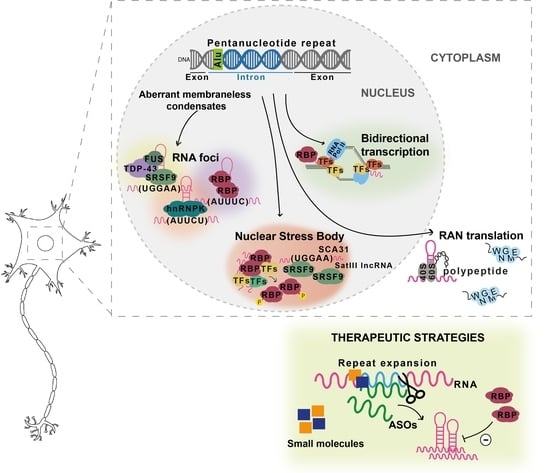
:1. Introduction
2. Pentanucleotide Repeats in Spinocerebellar Ataxias
2.1. ATXN10 ATTCT Repeat Expansions and Inserted Interruptions in SCA10
2.2. TGGAA Repeat Insertion in BEAN1 and TK2 in SCA31
2.3. DAB1 ATTTC Repeat Insertion in SCA37
3. Biallelic RFC1 AAGGG Expansions in CANVAS
4. Pentanucleotide Repeats in Familial Adult Myoclonic Epilepsy
4.1. SAMD12 Repeat Insertion in FAME1
4.2. STARD7 ATTTC Repeat Insertion in FAME2
4.3. MARCH6 Repeat Insertion in FAME3
4.4. Repeat Insertions in Other Types of FAME
5. Alu Repeat Expansions and Insertions
6. Therapeutic Strategies
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malik, I.; Kelley, C.P.; Wang, E.T.; Todd, P.K. Author Correction: Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 2021, 22, 644. [Google Scholar] [CrossRef] [PubMed]
- Morriss, G.R.; Cooper, T.A. Protein sequestration as a normal function of long noncoding RNAs and a pathogenic mechanism of RNAs containing nucleotide repeat expansions. Hum. Genet. 2017, 136, 1247–1263. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Mackowiak, S.D.; Niskanen, H.; Knezevic, D.; Asimi, V.; Grosswendt, S.; Geertsema, H.; Ali, S.; Jerkovic, I.; Ewers, H.; et al. Unblending of Transcriptional Condensates in Human Repeat Expansion Disease. Cell 2020, 181, 1062–1079.e1030. [Google Scholar] [CrossRef]
- Paul, S.; Dansithong, W.; Figueroa, K.P.; Scoles, D.R.; Pulst, S.M. Staufen1 links RNA stress granules and autophagy in a model of neurodegeneration. Nat. Commun. 2018, 9, 3648. [Google Scholar] [CrossRef]
- Loureiro, J.R.; Oliveira, C.L.; Silveira, I. Unstable repeat expansions in neurodegenerative diseases: Nucleocytoplasmic transport emerges on the scene. Neurobiol. Aging 2016, 39, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Cleary, J.D.; Ranum, L.P.W. Repeat-Associated Non-ATG Translation: Molecular Mechanisms and Contribution to Neurological Disease. Annu. Rev. Neurosci. 2019, 42, 227–247. [Google Scholar] [CrossRef]
- Sznajder, L.J.; Swanson, M.S. Short Tandem Repeat Expansions and RNA-Mediated Pathogenesis in Myotonic Dystrophy. Int. J. Mol. Sci. 2019, 20, 3365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalioti, M.D.; Scott, H.S.; Buresi, C.; Rossier, C.; Bottani, A.; Morris, M.A.; Malafosse, A.; Antonarakis, S.E. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature 1997, 386, 847–851. [Google Scholar] [CrossRef]
- LaCroix, A.J.; Stabley, D.; Sahraoui, R.; Adam, M.P.; Mehaffey, M.; Kernan, K.; Myers, C.T.; Fagerstrom, C.; Anadiotis, G.; Akkari, Y.M.; et al. GGC Repeat Expansion and Exon 1 Methylation of XYLT1 Is a Common Pathogenic Variant in Baratela-Scott Syndrome. Am. J. Hum. Genet. 2019, 104, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, S.E.; O’Hearn, E.E.; McInnis, M.G.; Gorelick-Feldman, D.A.; Kleiderlein, J.J.; Callahan, C.; Kwak, N.G.; Ingersoll-Ashworth, R.G.; Sherr, M.; Sumner, A.J.; et al. Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat. Genet. 1999, 23, 391–392. [Google Scholar] [CrossRef]
- O’Hearn, E.E.; Hwang, H.S.; Holmes, S.E.; Rudnicki, D.D.; Chung, D.W.; Seixas, A.I.; Cohen, R.L.; Ross, C.A.; Trojanowski, J.Q.; Pletnikova, O.; et al. Neuropathology and Cellular Pathogenesis of Spinocerebellar Ataxia Type 12. Mov. Disord. 2015, 30, 1813–1824. [Google Scholar] [CrossRef] [Green Version]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.P.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef]
- Knight, S.J.; Flannery, A.V.; Hirst, M.C.; Campbell, L.; Christodoulou, Z.; Phelps, S.R.; Pointon, J.; Middleton-Price, H.R.; Barnicoat, A.; Pembrey, M.E.; et al. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell 1993, 74, 127–134. [Google Scholar] [CrossRef]
- Metsu, S.; Rooms, L.; Rainger, J.; Taylor, M.S.; Bengani, H.; Wilson, D.I.; Chilamakuri, C.S.; Morrison, H.; Vandeweyer, G.; Reyniers, E.; et al. FRA2A is a CGG repeat expansion associated with silencing of AFF3. PLoS Genet. 2014, 10, e1004242. [Google Scholar] [CrossRef]
- Metsu, S.; Rainger, J.K.; Debacker, K.; Bernhard, B.; Rooms, L.; Grafodatskaya, D.; Weksberg, R.; Fombonne, E.; Taylor, M.S.; Scherer, S.W.; et al. A CGG-repeat expansion mutation in ZNF713 causes FRA7A: Association with autistic spectrum disorder in two families. Hum. Mutat. 2014, 35, 1295–1300. [Google Scholar] [CrossRef]
- Debacker, K.; Winnepenninckx, B.; Longman, C.; Colgan, J.; Tolmie, J.; Murray, R.; van Luijk, R.; Scheers, S.; Fitzpatrick, D.; Kooy, F. The molecular basis of the folate-sensitive fragile site FRA11A at 11q13. Cytogenet. Genome Res. 2007, 119, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Winnepenninckx, B.; Debacker, K.; Ramsay, J.; Smeets, D.; Smits, A.; FitzPatrick, D.R.; Kooy, R.F. CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am. J. Hum. Genet. 2007, 80, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Van Kuilenburg, A.B.P.; Tarailo-Graovac, M.; Richmond, P.A.; Drogemoller, B.I.; Pouladi, M.A.; Leen, R.; Brand-Arzamendi, K.; Dobritzsch, D.; Dolzhenko, E.; Eberle, M.A.; et al. Glutaminase Deficiency Caused by Short Tandem Repeat Expansion in GLS. N. Engl. J. Med. 2019, 380, 1433–1441. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Leehey, M.; Heinrichs, W.; Tassone, F.; Wilson, R.; Hills, J.; Grigsby, J.; Gage, B.; Hagerman, P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 2001, 57, 127–130. [Google Scholar] [CrossRef]
- Cronister, A.; Schreiner, R.; Wittenberger, M.; Amiri, K.; Harris, K.; Hagerman, R.J. Heterozygous fragile X female: Historical, physical, cognitive, and cytogenetic features. Am. J. Med. Genet. 1991, 38, 269–274. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, J.L.; Huang, W.; Zeng, S.; Jiao, B.; Liu, Z.; Chen, Z.; Li, Y.; Wang, Y.; Min, H.X.; et al. Expansion of Human-Specific GGC Repeat in Neuronal Intranuclear Inclusion Disease-Related Disorders. Am. J. Hum. Genet. 2019, 105, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Sone, J.; Mitsuhashi, S.; Fujita, A.; Mizuguchi, T.; Hamanaka, K.; Mori, K.; Koike, H.; Hashiguchi, A.; Takashima, H.; Sugiyama, H.; et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 2019, 51, 1215–1221. [Google Scholar] [CrossRef]
- Ishiura, H.; Shibata, S.; Yoshimura, J.; Suzuki, Y.; Qu, W.; Doi, K.; Almansour, M.A.; Kikuchi, J.K.; Taira, M.; Mitsui, J.; et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet. 2019, 51, 1222–1232. [Google Scholar] [CrossRef]
- Campuzano, V.; Montermini, L.; Molto, M.D.; Pianese, L.; Cossee, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996, 271, 1423–1427. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Renton, A.E.; Majounie, E.; Waite, A.; Simon-Sanchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, T.; Yamagata, T.; Burgess, D.L.; Rasmussen, A.; Grewal, R.P.; Watase, K.; Khajavi, M.; McCall, A.E.; Davis, C.F.; Zu, L.; et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000, 26, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murias, M.; Quintans, B.; Arias, M.; Seixas, A.I.; Cacheiro, P.; Tarrio, R.; Pardo, J.; Millan, M.J.; Arias-Rivas, S.; Blanco-Arias, P.; et al. ‘Costa da Morte’ ataxia is spinocerebellar ataxia 36: Clinical and genetic characterization. Brain 2012, 135, 1423–1435. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, H.; Abe, K.; Matsuura, T.; Ikeda, Y.; Hitomi, T.; Akechi, Y.; Habu, T.; Liu, W.; Okuda, H.; Koizumi, A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 2011, 89, 121–130. [Google Scholar] [CrossRef] [Green Version]
- Liquori, C.L.; Ricker, K.; Moseley, M.L.; Jacobsen, J.F.; Kress, W.; Naylor, S.L.; Day, J.W.; Ranum, L.P. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 2001, 293, 864–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieben, E.D.; Aleff, R.A.; Tosakulwong, N.; Butz, M.L.; Highsmith, W.E.; Edwards, A.O.; Baratz, K.H. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS ONE 2012, 7, e49083. [Google Scholar] [CrossRef]
- Cortese, A.; Simone, R.; Sullivan, R.; Vandrovcova, J.; Tariq, H.; Yau, W.Y.; Humphrey, J.; Jaunmuktane, Z.; Sivakumar, P.; Polke, J.; et al. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat. Genet. 2019, 51, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Rafehi, H.; Szmulewicz, D.J.; Bennett, M.F.; Sobreira, N.L.M.; Pope, K.; Smith, K.R.; Gillies, G.; Diakumis, P.; Dolzhenko, E.; Eberle, M.A.; et al. Bioinformatics-Based Identification of Expanded Repeats: A Non-reference Intronic Pentamer Expansion in RFC1 Causes CANVAS. Am. J. Hum. Genet. 2019, 105, 151–165. [Google Scholar] [CrossRef] [Green Version]
- Sato, N.; Amino, T.; Kobayashi, K.; Asakawa, S.; Ishiguro, T.; Tsunemi, T.; Takahashi, M.; Matsuura, T.; Flanigan, K.M.; Iwasaki, S.; et al. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am. J. Hum. Genet. 2009, 85, 544–557. [Google Scholar] [CrossRef] [Green Version]
- Seixas, A.I.; Loureiro, J.R.; Costa, C.; Ordonez-Ugalde, A.; Marcelino, H.; Oliveira, C.L.; Loureiro, J.L.; Dhingra, A.; Brandao, E.; Cruz, V.T.; et al. A Pentanucleotide ATTTC Repeat Insertion in the Non-coding Region of DAB1, Mapping to SCA37, Causes Spinocerebellar Ataxia. Am. J. Hum. Genet. 2017, 101, 87–103. [Google Scholar] [CrossRef] [Green Version]
- Ishiura, H.; Doi, K.; Mitsui, J.; Yoshimura, J.; Matsukawa, M.K.; Fujiyama, A.; Toyoshima, Y.; Kakita, A.; Takahashi, H.; Suzuki, Y.; et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 2018, 50, 581–590. [Google Scholar] [CrossRef]
- Corbett, M.A.; Kroes, T.; Veneziano, L.; Bennett, M.F.; Florian, R.; Schneider, A.L.; Coppola, A.; Licchetta, L.; Franceschetti, S.; Suppa, A.; et al. Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. Nat. Commun. 2019, 10, 4920. [Google Scholar] [CrossRef] [Green Version]
- Florian, R.T.; Kraft, F.; Leitao, E.; Kaya, S.; Klebe, S.; Magnin, E.; van Rootselaar, A.F.; Buratti, J.; Kuhnel, T.; Schroder, C.; et al. Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3. Nat. Commun. 2019, 10, 4919. [Google Scholar] [CrossRef]
- Yeetong, P.; Pongpanich, M.; Srichomthong, C.; Assawapitaksakul, A.; Shotelersuk, V.; Tantirukdham, N.; Chunharas, C.; Suphapeetiporn, K.; Shotelersuk, V. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain 2019, 142, 3360–3366. [Google Scholar] [CrossRef]
- Bragg, D.C.; Mangkalaphiban, K.; Vaine, C.A.; Kulkarni, N.J.; Shin, D.; Yadav, R.; Dhakal, J.; Ton, M.L.; Cheng, A.; Russo, C.T.; et al. Disease onset in X-linked dystonia-parkinsonism correlates with expansion of a hexameric repeat within an SVA retrotransposon in TAF1. Proc. Natl. Acad. Sci. USA 2017, 114, E11020–E11028. [Google Scholar] [CrossRef] [Green Version]
- Mahadevan, M.; Tsilfidis, C.; Sabourin, L.; Shutler, G.; Amemiya, C.; Jansen, G.; Neville, C.; Narang, M.; Barcelo, J.; O’Hoy, K.; et al. Myotonic dystrophy mutation: An unstable CTG repeat in the 3′ untranslated region of the gene. Science 1992, 255, 1253–1255. [Google Scholar] [CrossRef]
- Brook, J.D.; McCurrach, M.E.; Harley, H.G.; Buckler, A.J.; Church, D.; Aburatani, H.; Hunter, K.; Stanton, V.P.; Thirion, J.P.; Hudson, T.; et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 1992, 68, 799–808. [Google Scholar] [CrossRef]
- Seixas, A.I.; Holmes, S.E.; Takeshima, H.; Pavlovich, A.; Sachs, N.; Pruitt, J.L.; Silveira, I.; Ross, C.A.; Margolis, R.L.; Rudnicki, D.D. Loss of junctophilin-3 contributes to huntington disease-like 2 pathogenesis. Ann. Neurol. 2012, 71, 245–257. [Google Scholar] [CrossRef]
- Wilburn, B.; Rudnicki, D.D.; Zhao, J.; Weitz, T.M.; Cheng, Y.; Gu, X.; Greiner, E.; Park, C.S.; Wang, N.; Sopher, B.L.; et al. An antisense CAG repeat transcript at JPH3 locus med.diates expanded polyglutamine protein toxicity in Huntington’s disease-like 2 mice. Neuron 2011, 70, 427–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, S.E.; O’Hearn, E.; Rosenblatt, A.; Callahan, C.; Hwang, H.S.; Ingersoll-Ashworth, R.G.; Fleisher, A.; Stevanin, G.; Brice, A.; Potter, N.T.; et al. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat. Genet. 2001, 29, 377–378. [Google Scholar] [CrossRef]
- Koob, M.D.; Moseley, M.L.; Schut, L.J.; Benzow, K.A.; Bird, T.D.; Day, J.W.; Ranum, L.P. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat. Genet. 1999, 21, 379–384. [Google Scholar] [CrossRef]
- Moseley, M.L.; Zu, T.; Ikeda, Y.; Gao, W.; Mosemiller, A.K.; Daughters, R.S.; Chen, G.; Weatherspoon, M.R.; Clark, H.B.; Ebner, T.J.; et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006, 38, 758–769. [Google Scholar] [CrossRef]
- Koide, R.; Ikeuchi, T.; Onodera, O.; Tanaka, H.; Igarashi, S.; Endo, K.; Takahashi, H.; Kondo, R.; Ishikawa, A.; Hayashi, T.; et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA). Nat. Genet. 1994, 6, 9–13. [Google Scholar] [CrossRef]
- The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Orr, H.T.; Chung, M.Y.; Banfi, S.; Kwiatkowski, T.J.; Servadio, A.; Beaudet, A.L.; McCall, A.E.; Duvick, L.A.; Ranum, L.P.; Zoghbi, H.Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 1993, 4, 221–226. [Google Scholar] [CrossRef]
- La Spada, A.R.; Wilson, E.M.; Lubahn, D.B.; Harding, A.E.; Fischbeck, K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 1991, 352, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Imbert, G.; Saudou, F.; Yvert, G.; Devys, D.; Trottier, Y.; Garnier, J.M.; Weber, C.; Mandel, J.L.; Cancel, G.; Abbas, N.; et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 1996, 14, 285–291. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Okamoto, T.; Taniwaki, M.; Aizawa, M.; Inoue, M.; Katayama, S.; Kawakami, H.; Nakamura, S.; Nishimura, M.; Akiguchi, I. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994, 8, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Zhuchenko, O.; Bailey, J.; Bonnen, P.; Ashizawa, T.; Stockton, D.W.; Amos, C.; Dobyns, W.B.; Subramony, S.H.; Zoghbi, H.Y.; Lee, C.C. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat. Genet. 1997, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Lindblad, K.; Savontaus, M.L.; Stevanin, G.; Holmberg, M.; Digre, K.; Zander, C.; Ehrsson, H.; David, G.; Benomar, A.; Nikoskelainen, E.; et al. An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res. 1996, 6, 965–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koide, R.; Kobayashi, S.; Shimohata, T.; Ikeuchi, T.; Maruyama, M.; Saito, M.; Yamada, M.; Takahashi, H.; Tsuji, S. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: A new polyglutamine disease? Hum. Mol. Genet. 1999, 8, 2047–2053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brais, B.; Bouchard, J.P.; Xie, Y.G.; Rochefort, D.L.; Chretien, N.; Tome, F.M.; Lafreniere, R.G.; Rommens, J.M.; Uyama, E.; Nohira, O.; et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 1998, 18, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Stromme, P.; Mangelsdorf, M.E.; Shaw, M.A.; Lower, K.M.; Lewis, S.M.; Bruyere, H.; Lutcherath, V.; Gedeon, A.K.; Wallace, R.H.; Scheffer, I.E.; et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat. Genet. 2002, 30, 441–445. [Google Scholar] [CrossRef]
- De Baere, E.; Beysen, D.; Oley, C.; Lorenz, B.; Cocquet, J.; De Sutter, P.; Devriendt, K.; Dixon, M.; Fellous, M.; Fryns, J.P.; et al. FOXL2 and BPES: Mutational hotspots, phenotypic variability, and revision of the genotype-phenotype correlation. Am. J. Hum. Genet. 2003, 72, 478–487. [Google Scholar] [CrossRef] [Green Version]
- Mundlos, S.; Otto, F.; Mundlos, C.; Mulliken, J.B.; Aylsworth, A.S.; Albright, S.; Lindhout, D.; Cole, W.G.; Henn, W.; Knoll, J.H.; et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997, 89, 773–779. [Google Scholar] [CrossRef] [Green Version]
- Amiel, J.; Laudier, B.; Attie-Bitach, T.; Trang, H.; de Pontual, L.; Gener, B.; Trochet, D.; Etchevers, H.; Ray, P.; Simonneau, M.; et al. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet. 2003, 33, 459–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, F.R.; Bacchelli, C.; Brady, A.F.; Brueton, L.A.; Fryns, J.P.; Mortlock, D.P.; Innis, J.W.; Holmes, L.B.; Donnenfeld, A.E.; Feingold, M.; et al. Novel HOXA13 mutations and the phenotypic spectrum of hand-foot-genital syndrome. Am. J. Hum. Genet. 2000, 67, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.A.; Warburton, D.; Brown, L.Y.; Yu, C.Y.; Roeder, E.R.; Stengel-Rutkowski, S.; Hennekam, R.C.; Muenke, M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 1998, 20, 180–183. [Google Scholar] [CrossRef]
- Muragaki, Y.; Mundlos, S.; Upton, J.; Olsen, B.R. Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 1996, 272, 548–551. [Google Scholar] [CrossRef]
- Kato, M.; Das, S.; Petras, K.; Kitamura, K.; Morohashi, K.; Abuelo, D.N.; Barr, M.; Bonneau, D.; Brady, A.F.; Carpenter, N.J.; et al. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum. Mutat. 2004, 23, 147–159. [Google Scholar] [CrossRef]
- Laumonnier, F.; Ronce, N.; Hamel, B.C.; Thomas, P.; Lespinasse, J.; Raynaud, M.; Paringaux, C.; Van Bokhoven, H.; Kalscheuer, V.; Fryns, J.P.; et al. Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am. J. Hum. Genet. 2002, 71, 1450–1455. [Google Scholar] [CrossRef] [Green Version]
- McLoughlin, H.S.; Moore, L.R.; Paulson, H.L. Pathogenesis of SCA3 and implications for other polyglutamine diseases. Neurobiol. Dis. 2019, 134, 104635. [Google Scholar] [CrossRef]
- Abu-Baker, A.; Rouleau, G.A. Oculopharyngeal muscular dystrophy: Recent advances in the understanding of the molecular pathogenic mechanisms and treatment strategies. Biochim. Biophys. Acta 2007, 1772, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.N.; Thomas, P.Q. Molecular pathology of polyalanine expansion disorders: New perspectives from mouse models. Methods Mol. Biol. 2013, 1017, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Massey, T.H.; Jones, L. The central role of DNA damage and repair in CAG repeat diseases. Dis. Model. Mech. 2018, 11, dmm031930. [Google Scholar] [CrossRef] [Green Version]
- Quinodoz, S.A.; Jachowicz, J.W.; Bhat, P.; Ollikainen, N.; Banerjee, A.K.; Goronzy, I.N.; Blanco, M.R.; Chovanec, P.; Chow, A.; Markaki, Y.; et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 2021, 184, 5775–5790.e30. [Google Scholar] [CrossRef]
- Wiedner, H.J.; Giudice, J. It’s not just a phase: Function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biol. 2021, 28, 465–473. [Google Scholar] [CrossRef]
- Nelson, D.L.; Orr, H.T.; Warren, S.T. The unstable repeats—Three evolving faces of neurological disease. Neuron 2013, 77, 825–843. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lu, Y.; Polak, U.; Lin, K.; Shen, J.; Farmer, J.; Seyer, L.; Bhalla, A.D.; Rozwadowska, N.; Lynch, D.R.; et al. Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus. Hum. Mol. Genet. 2015, 24, 6932–6943. [Google Scholar] [CrossRef] [Green Version]
- Mankodi, A.; Logigian, E.; Callahan, L.; McClain, C.; White, R.; Henderson, D.; Krym, M.; Thornton, C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 2000, 289, 1769–1773. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Li, M.; Manchanda, M.; Batra, R.; Charizanis, K.; Mohan, A.; Warren, S.A.; Chamberlain, C.M.; Finn, D.; Hong, H.; et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 2013, 5, 1887–1900. [Google Scholar] [CrossRef]
- Jain, A.; Vale, R.D. RNA phase transitions in repeat expansion disorders. Nature 2017, 546, 243–247. [Google Scholar] [CrossRef] [Green Version]
- Hall, L.L.; Lawrence, J.B. RNA as a fundamental component of interphase chromosomes: Could repeats prove key? Curr. Opin. Genet. Dev. 2016, 37, 137–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zu, T.; Gibbens, B.; Doty, N.S.; Gomes-Pereira, M.; Huguet, A.; Stone, M.D.; Margolis, J.; Peterson, M.; Markowski, T.W.; Ingram, M.A.; et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA 2011, 108, 260–265. [Google Scholar] [CrossRef] [Green Version]
- Cleary, J.D.; Pattamatta, A.; Ranum, L.P.W. Repeat-associated non-ATG (RAN) translation. J. Biol. Chem. 2018, 293, 16127–16141. [Google Scholar] [CrossRef] [Green Version]
- Cho, D.H.; Thienes, C.P.; Mahoney, S.E.; Analau, E.; Filippova, G.N.; Tapscott, S.J. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell 2005, 20, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Ladd, P.D.; Smith, L.E.; Rabaia, N.A.; Moore, J.M.; Georges, S.A.; Hansen, R.S.; Hagerman, R.J.; Tassone, F.; Tapscott, S.J.; Filippova, G.N. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007, 16, 3174–3187. [Google Scholar] [CrossRef] [Green Version]
- Zu, T.; Liu, Y.; Banez-Coronel, M.; Reid, T.; Pletnikova, O.; Lewis, J.; Miller, T.M.; Harms, M.B.; Falchook, A.E.; Subramony, S.H.; et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA 2013, 110, E4968–E4977. [Google Scholar] [CrossRef] [Green Version]
- Castro, A.F.; Loureiro, J.R.; Bessa, J.; Silveira, I. Antisense Transcription across Nucleotide Repeat Expansions in Neurodegenerative and Neuromuscular Diseases: Progress and Mysteries. Genes 2020, 11, 1418. [Google Scholar] [CrossRef]
- Plys, A.J.; Kingston, R.E. Dynamic condensates activate transcription. Science 2018, 361, 329–330. [Google Scholar] [CrossRef]
- Ishikawa, K.; Nagai, Y. Molecular Mechanisms and Future Therapeutics for Spinocerebellar Ataxia Type 31 (SCA31). Neurotherapeutics 2019, 16, 1106–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loureiro, J.R.; Oliveira, C.L.; Mota, C.; Castro, A.F.; Costa, C.; Loureiro, J.L.; Coutinho, P.; Martins, S.; Sequeiros, J.; Silveira, I. Mutational mechanism for DAB1 (ATTTC)n insertion in SCA37: ATTTT repeat lengthening and nucleotide substitution. Hum. Mutat. 2019, 40, 404–412. [Google Scholar] [CrossRef]
- Klockgether, T.; Mariotti, C.; Paulson, H.L. Spinocerebellar ataxia. Nat. Rev. Dis. Primers 2019, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Verbeek, D.S. Why do so many genetic insults lead to Purkinje Cell degeneration and spinocerebellar ataxia? Neurosci. Lett. 2019, 688, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Buijsen, R.A.M.; Toonen, L.J.A.; Gardiner, S.L.; van Roon-Mom, W.M.C. Genetics, Mechanisms, and Therapeutic Progress in Polyglutamine Spinocerebellar Ataxias. Neurotherapeutics 2019, 16, 263–286. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Ashizawa, T. SCA10 and ATTCT repeat expansion: Clinical features and molecular aspects. Cytogenet. Genome Res. 2003, 100, 184–188. [Google Scholar] [CrossRef]
- Teive, H.A.; Roa, B.B.; Raskin, S.; Fang, P.; Arruda, W.O.; Neto, Y.C.; Gao, R.; Werneck, L.C.; Ashizawa, T. Clinical phenotype of Brazilian families with spinocerebellar ataxia 10. Neurology 2004, 63, 1509–1512. [Google Scholar] [CrossRef]
- Alonso, I.; Jardim, L.; Artigalas, O.; Saraiva-Pereira, M.; Matsuura, T.; Ashizawa, T.; Sequeiros, J.; Silveira, I. Reduced penetrance of intermediate size alleles in spinocerebellar ataxia type 10. Neurology 2006, 66, 1602–1604. [Google Scholar] [CrossRef]
- Rasmussen, A.; Matsuura, T.; Ruano, L.; Yescas, P.; Ochoa, A.; Ashizawa, T.; Alonso, E. Clinical and genetic analysis of four Mexican families with spinocerebellar ataxia type 10. Ann. Neurol. 2001, 50, 234–239. [Google Scholar] [CrossRef]
- Almeida, T.; Alonso, I.; Martins, S.; Ramos, E.M.; Azevedo, L.; Ohno, K.; Amorim, A.; Saraiva-Pereira, M.L.; Jardim, L.B.; Matsuura, T.; et al. Ancestral origin of the ATTCT repeat expansion in spinocerebellar ataxia type 10 (SCA10). PLoS ONE 2009, 4, e4553. [Google Scholar] [CrossRef] [Green Version]
- Bushara, K.; Bower, M.; Liu, J.; McFarland, K.N.; Landrian, I.; Hutter, D.; Teive, H.A.; Rasmussen, A.; Mulligan, C.J.; Ashizawa, T. Expansion of the Spinocerebellar ataxia type 10 (SCA10) repeat in a patient with Sioux Native American ancestry. PLoS ONE 2013, 8, e81342. [Google Scholar] [CrossRef] [Green Version]
- Leonardi, L.; Marcotulli, C.; McFarland, K.N.; Tessa, A.; DiFabio, R.; Santorelli, F.M.; Pierelli, F.; Ashizawa, T.; Casali, C. Spinocerebellar ataxia type 10 in Peru: The missing link in the Amerindian origin of the disease. J. Neurol. 2014, 261, 1691–1694. [Google Scholar] [CrossRef] [Green Version]
- Baizabal-Carvallo, J.F.; Xia, G.; Botros, P.; Laguna, J.; Ashizawa, T.; Jankovic, J. Bolivian kindred with combined spinocerebellar ataxia types 2 and 10. Acta Neurol. Scand. 2015, 132, 139–142. [Google Scholar] [CrossRef] [Green Version]
- Cornejo-Olivas, M.; Inca-Martinez, M.; Castilhos, R.M.; Furtado, G.V.; Mattos, E.P.; Bampi, G.B.; Leistner-Segal, S.; Marca, V.; Mazzetti, P.; Saraiva-Pereira, M.L.; et al. Genetic Analysis of Hereditary Ataxias in Peru Identifies SCA10 Families with Incomplete Penetrance. Cerebellum 2020, 19, 208–215. [Google Scholar] [CrossRef]
- Wang, K.; McFarland, K.N.; Liu, J.; Zeng, D.; Landrian, I.; Xia, G.; Hao, Y.; Jin, M.; Mulligan, C.J.; Gu, W.; et al. Spinocerebellar ataxia type 10 in Chinese Han. Neurol. Genet. 2015, 1, e26. [Google Scholar] [CrossRef]
- Naito, H.; Takahashi, T.; Kamada, M.; Morino, H.; Yoshino, H.; Hattori, N.; Maruyama, H.; Kawakami, H.; Matsumoto, M. First report of a Japanese family with spinocerebellar ataxia type 10: The second report from Asia after a report from China. PLoS ONE 2017, 12, e0177955. [Google Scholar] [CrossRef] [PubMed]
- Waragai, M.; Nagamitsu, S.; Xu, W.; Li, Y.J.; Lin, X.; Ashizawa, T. Ataxin 10 induces neuritogenesis via interaction with G-protein beta2 subunit. J. Neurosci. Res. 2006, 83, 1170–1178. [Google Scholar] [CrossRef]
- Matsuura, T.; Fang, P.; Pearson, C.E.; Jayakar, P.; Ashizawa, T.; Roa, B.B.; Nelson, D.L. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: Repeat purity as a disease modifier? Am. J. Hum. Genet. 2006, 78, 125–129. [Google Scholar] [CrossRef] [Green Version]
- McFarland, K.N.; Liu, J.; Landrian, I.; Godiska, R.; Shanker, S.; Yu, F.; Farmerie, W.G.; Ashizawa, T. SMRT Sequencing of Long Tandem Nucleotide Repeats in SCA10 Reveals Unique Insight of Repeat Expansion Structure. PLoS ONE 2015, 10, e0135906. [Google Scholar] [CrossRef]
- McFarland, K.N.; Liu, J.; Landrian, I.; Zeng, D.; Raskin, S.; Moscovich, M.; Gatto, E.M.; Ochoa, A.; Teive, H.A.; Rasmussen, A.; et al. Repeat interruptions in spinocerebellar ataxia type 10 expansions are strongly associated with epileptic seizures. Neurogenetics 2014, 15, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Hashem, V.; Tiwari, A.; Bewick, B.; Teive, H.A.G.; Moscovich, M.; Schuele, B.; Bushara, K.; Bower, M.; Rasmussen, A.; Tsai, Y.C.; et al. Pulse-Field capillary electrophoresis of repeat-primed PCR amplicons for analysis of large repeats in Spinocerebellar Ataxia Type 10. PLoS ONE 2020, 15, e0228789. [Google Scholar] [CrossRef]
- Kurosaki, T.; Matsuura, T.; Ohno, K.; Ueda, S. Alu-mediated acquisition of unstable ATTCT pentanucleotide repeats in the human ATXN10 gene. Mol. Biol. Evol. 2009, 26, 2573–2579. [Google Scholar] [CrossRef]
- Wakamiya, M.; Matsuura, T.; Liu, Y.; Schuster, G.C.; Gao, R.; Xu, W.; Sarkar, P.S.; Lin, X.; Ashizawa, T. The role of ataxin 10 in the pathogenesis of spinocerebellar ataxia type 10. Neurology 2006, 67, 607–613. [Google Scholar] [CrossRef]
- White, M.C.; Gao, R.; Xu, W.; Mandal, S.M.; Lim, J.G.; Hazra, T.K.; Wakamiya, M.; Edwards, S.F.; Raskin, S.; Teive, H.A.; et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010, 6, e1000984. [Google Scholar] [CrossRef]
- White, M.; Xia, G.; Gao, R.; Wakamiya, M.; Sarkar, P.S.; McFarland, K.; Ashizawa, T. Transgenic mice with SCA10 pentanucleotide repeats show motor phenotype and susceptibility to seizure: A toxic RNA gain-of-function model. J. Neurosci. Res. 2012, 90, 706–714. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, K.; Durr, A.; Klopstock, T.; Muller, S.; De Toffol, B.; Vidailhet, M.; Vighetto, A.; Marelli, C.; Wichmann, H.E.; Illig, T.; et al. Pentanucleotide repeats at the spinocerebellar ataxia type 31 (SCA31) locus in Caucasians. Neurology 2011, 77, 1853–1855. [Google Scholar] [CrossRef]
- Sakai, H.; Yoshida, K.; Shimizu, Y.; Morita, H.; Ikeda, S.; Matsumoto, N. Analysis of an insertion mutation in a cohort of 94 patients with spinocerebellar ataxia type 31 from Nagano, Japan. Neurogenetics 2010, 11, 409–415. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Matsushima, A.; Nakamura, K. Inter-generational instability of inserted repeats during transmission in spinocerebellar ataxia type 31. J. Hum. Genet. 2017, 62, 923–925. [Google Scholar] [CrossRef]
- Lee, Y.C.; Liu, C.S.; Lee, T.Y.; Lo, Y.C.; Lu, Y.C.; Soong, B.W. SCA31 is rare in the Chinese population on Taiwan. Neurobiol. Aging 2012, 33, 426.e23–426.e24. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; He, Z.; Li, L.; Qin, X.; Zhao, Y.; Yuan, L. Spinocerebellar ataxia type 31 exists in northeast China. J. Neurol. Sci. 2012, 316, 164–167. [Google Scholar] [CrossRef]
- Niimi, Y.; Takahashi, M.; Sugawara, E.; Umeda, S.; Obayashi, M.; Sato, N.; Ishiguro, T.; Higashi, M.; Eishi, Y.; Mizusawa, H.; et al. Abnormal RNA structures (RNA foci) containing a penta-nucleotide repeat (UGGAA)n in the Purkinje cell nucleus is associated with spinocerebellar ataxia type 31 pathogenesis. Neuropathology 2013, 33, 600–611. [Google Scholar] [CrossRef]
- Ishiguro, T.; Sato, N.; Ueyama, M.; Fujikake, N.; Sellier, C.; Kanegami, A.; Tokuda, E.; Zamiri, B.; Gall-Duncan, T.; Mirceta, M.; et al. Regulatory Role of RNA Chaperone TDP-43 for RNA Misfolding and Repeat-Associated Translation in SCA31. Neuron 2017, 94, 108–124.e7. [Google Scholar] [CrossRef] [Green Version]
- Shibata, T.; Nagano, K.; Ueyama, M.; Ninomiya, K.; Hirose, T.; Nagai, Y.; Ishikawa, K.; Kawai, G.; Nakatani, K. Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model. Nat. Commun. 2021, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, K.; Adachi, S.; Natsume, T.; Iwakiri, J.; Terai, G.; Asai, K.; Hirose, T. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 2020, 39, e102729. [Google Scholar] [CrossRef]
- Loureiro, J.R.; Oliveira, C.L.; Sequeiros, J.; Silveira, I. A repeat-primed PCR assay for pentanucleotide repeat alleles in spinocerebellar ataxia type 37. J. Hum. Genet. 2018, 63, 981–987. [Google Scholar] [CrossRef]
- Serrano-Munuera, C.; Corral-Juan, M.; Stevanin, G.; San Nicolas, H.; Roig, C.; Corral, J.; Campos, B.; de Jorge, L.; Morcillo-Suarez, C.; Navarro, A.; et al. New subtype of spinocerebellar ataxia with altered vertical eye movements mapping to chromosome 1p32. JAMA Neurol. 2013, 70, 764–771. [Google Scholar] [CrossRef] [Green Version]
- Corral-Juan, M.; Serrano-Munuera, C.; Rabano, A.; Cota-Gonzalez, D.; Segarra-Roca, A.; Ispierto, L.; Cano-Orgaz, A.T.; Adarmes, A.D.; Mendez-Del-Barrio, C.; Jesus, S.; et al. Clinical, genetic and neuropathological characterization of spinocerebellar ataxia type 37. Brain 2018, 141, 1981–1997. [Google Scholar] [CrossRef]
- Chung, M.Y.; Ranum, L.P.; Duvick, L.A.; Servadio, A.; Zoghbi, H.Y.; Orr, H.T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat. Genet. 1993, 5, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.M.; Martins, S.; Alonso, I.; Emmel, V.E.; Saraiva-Pereira, M.L.; Jardim, L.B.; Coutinho, P.; Sequeiros, J.; Silveira, I. Common Origin of Pure and Interrupted Repeat Expansions in Spinocerebellar Ataxia Type 2 (SCA2). Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010, 153B, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Liquori, C.L.; Ikeda, Y.; Weatherspoon, M.; Ricker, K.; Schoser, B.G.; Dalton, J.C.; Day, J.W.; Ranum, L.P. Myotonic dystrophy type 2: Human founder haplotype and evolutionary conservation of the repeat tract. Am. J. Hum. Genet. 2003, 73, 849–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daughters, R.S.; Tuttle, D.L.; Gao, W.; Ikeda, Y.; Moseley, M.L.; Ebner, T.J.; Swanson, M.S.; Ranum, L.P. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009, 5, e1000600. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, M.; Rice, D.S.; D’Arcangelo, G.; Yoneshima, H.; Nakajima, K.; Mikoshiba, K.; Howell, B.W.; Cooper, J.A.; Goldowitz, D.; Curran, T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature 1997, 389, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Szmulewicz, D.J.; Roberts, L.; McLean, C.A.; MacDougall, H.G.; Halmagyi, G.M.; Storey, E. Proposed diagnostic criteria for cerebellar ataxia with neuropathy and vestibular areflexia syndrome (CANVAS). Neurol. Clin. Pract. 2016, 6, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akcimen, F.; Ross, J.P.; Bourassa, C.V.; Liao, C.; Rochefort, D.; Gama, M.T.D.; Dicaire, M.J.; Barsottini, O.G.; Brais, B.; Pedroso, J.L.; et al. Investigation of the RFC1 Repeat Expansion in a Canadian and a Brazilian Ataxia Cohort: Identification of Novel Conformations. Front. Genet. 2019, 10, 1219. [Google Scholar] [CrossRef] [Green Version]
- Scriba, C.K.; Beecroft, S.J.; Clayton, J.S.; Cortese, A.; Sullivan, R.; Yau, W.Y.; Dominik, N.; Rodrigues, M.; Walker, E.; Dyer, Z.; et al. A novel RFC1 repeat motif (ACAGG) in two Asia-Pacific CANVAS families. Brain 2020, 143, 2904–2910. [Google Scholar] [CrossRef] [PubMed]
- Beecroft, S.J.; Cortese, A.; Sullivan, R.; Yau, W.Y.; Dyer, Z.; Wu, T.Y.; Mulroy, E.; Pelosi, L.; Rodrigues, M.; Taylor, R.; et al. A Maori specific RFC1 pathogenic repeat configuration in CANVAS, likely due to a founder allele. Brain 2020, 143, 2673–2680. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Ueda, S.; Ishida, T.; Abe, K.; Ohno, K.; Matsuura, T. The unstable CCTG repeat responsible for myotonic dystrophy type 2 originates from an AluSx element insertion into an early primate genome. PLoS ONE 2012, 7, e38379. [Google Scholar] [CrossRef] [PubMed]
- Pastore, A.; Puccio, H. Frataxin: A protein in search for a function. J. Neurochem. 2013, 126 (Suppl. S1), 43–52. [Google Scholar] [CrossRef]
- Van den Ende, T.; Sharifi, S.; van der Salm, S.M.A.; van Rootselaar, A.F. Familial Cortical Myoclonic Tremor and Epilepsy, an Enigmatic Disorder: From Phenotypes to Pathophysiology and Genetics. A Systematic Review. Tremor Other Hyperkinet. Mov. 2018, 8, 503. [Google Scholar] [CrossRef]
- Depienne, C.; Magnin, E.; Bouteiller, D.; Stevanin, G.; Saint-Martin, C.; Vidailhet, M.; Apartis, E.; Hirsch, E.; LeGuern, E.; Labauge, P.; et al. Familial cortical myoclonic tremor with epilepsy: The third locus (FCMTE3) maps to 5p. Neurology 2010, 74, 2000–2003. [Google Scholar] [CrossRef] [PubMed]
- Striano, P.; Caranci, F.; Di Benedetto, R.; Tortora, F.; Zara, F.; Striano, S. 1H-MR spectroscopy indicates prominent cerebellar dysfunction in benign adult familial myoclonic epilepsy. Epilepsia 2009, 50, 1491–1497. [Google Scholar] [CrossRef]
- Van Rootselaar, A.F.; van der Salm, S.M.; Bour, L.J.; Edwards, M.J.; Brown, P.; Aronica, E.; Rozemuller-Kwakkel, J.M.; Koehler, P.J.; Koelman, J.H.; Rothwell, J.C.; et al. Decreased cortical inhibition and yet cerebellar pathology in ‘familial cortical myoclonic tremor with epilepsy’. Mov. Disord. 2007, 22, 2378–2385. [Google Scholar] [CrossRef]
- Buijink, A.W.; Broersma, M.; van der Stouwe, A.M.; Sharifi, S.; Tijssen, M.A.; Speelman, J.D.; Maurits, N.M.; van Rootselaar, A.F. Cerebellar Atrophy in Cortical Myoclonic Tremor and Not in Hereditary Essential Tremor-a Voxel-Based Morphometry Study. Cerebellum 2016, 15, 696–704. [Google Scholar] [CrossRef]
- Striano, P.; Louis, E.D.; Manto, M. Autosomal dominant cortical tremor, myoclonus, and epilepsy: Is the origin in the cerebellum? Editorial. Cerebellum 2013, 12, 145–146. [Google Scholar] [CrossRef] [Green Version]
- Depienne, C.; Mandel, J.L. 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? Am. J. Hum. Genet. 2021, 108, 764–785. [Google Scholar] [CrossRef] [PubMed]
- Mikami, M.; Yasuda, T.; Terao, A.; Nakamura, M.; Ueno, S.; Tanabe, H.; Tanaka, T.; Onuma, T.; Goto, Y.; Kaneko, S.; et al. Localization of a gene for benign adult familial myoclonic epilepsy to chromosome 8q23.3-q24.1. Am. J. Hum. Genet. 1999, 65, 745–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaster, N.M.; Uyama, E.; Uchino, M.; Ikeda, T.; Flanigan, K.M.; Kondo, I.; Ptacek, L.J. Genetic localization of the familial adult myoclonic epilepsy (FAME) gene to chromosome 8q24. Neurology 1999, 53, 1180–1183. [Google Scholar] [CrossRef] [PubMed]
- Cen, Z.; Jiang, Z.; Chen, Y.; Zheng, X.; Xie, F.; Yang, X.; Lu, X.; Ouyang, Z.; Wu, H.; Chen, S.; et al. Intronic pentanucleotide TTTCA repeat insertion in the SAMD12 gene causes familial cortical myoclonic tremor with epilepsy type 1. Brain 2018, 141, 2280–2288. [Google Scholar] [CrossRef] [PubMed]
- Cen, Z.; Chen, Y.; Yang, D.; Zhu, Q.; Chen, S.; Chen, X.; Wang, B.; Xie, F.; Ouyang, Z.; Jiang, Z.; et al. Intronic (TTTGA)n insertion in SAMD12 also causes familial cortical myoclonic tremor with epilepsy. Mov. Disord. 2019, 34, 1571–1576. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.X.; Liu, Q.; Lu, Q.; Huang, Y.; Zhou, X.Q.; Sun, H.Y.; Wu, L.W.; Cui, L.Y.; Zhang, X. TTTCA repeat expansion causes familial cortical myoclonic tremor with epilepsy. Eur. J. Neurol. 2019, 26, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Zhang, M.Y.; Wang, X.J.; Hu, Z.M.; Li, J.C.; Li, N.; Wang, J.L.; Liang, F.; Yang, Q.; Liu, Q.; et al. Long-read sequencing identified intronic repeat expansions in SAMD12 from Chinese pedigrees affected with familial cortical myoclonic tremor with epilepsy. J. Med. Genet. 2019, 56, 265–270. [Google Scholar] [CrossRef]
- Mizuguchi, T.; Toyota, T.; Adachi, H.; Miyake, N.; Matsumoto, N.; Miyatake, S. Detecting a long insertion variant in SAMD12 by SMRT sequencing: Implications of long-read whole-genome sequencing for repeat expansion diseases. J. Hum. Genet. 2019, 64, 191–197. [Google Scholar] [CrossRef]
- Bennett, M.F.; Oliver, K.L.; Regan, B.M.; Bellows, S.T.; Schneider, A.L.; Rafehi, H.; Sikta, N.; Crompton, D.E.; Coleman, M.; Hildebrand, M.S.; et al. Familial adult myoclonic epilepsy type 1 SAMD12 TTTCA repeat expansion arose 17,000 years ago and is present in Sri Lankan and Indian families. Eur. J. Hum. Genet. 2020, 28, 973–978. [Google Scholar] [CrossRef]
- Yeetong, P.; Chunharas, C.; Pongpanich, M.; Bennett, M.F.; Srichomthong, C.; Pasutharnchat, N.; Suphapeetiporn, K.; Bahlo, M.; Shotelersuk, V. Founder effect of the TTTCA repeat insertions in SAMD12 causing BAFME1. Eur. J. Hum. Genet. 2021, 29, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, R.; Bonanni, P.; Patrignani, A.; Brown, P.; Parmeggiani, L.; Grosse, P.; Brovedani, P.; Moro, F.; Aridon, P.; Carrozzo, R.; et al. Autosomal dominant cortical myoclonus and epilepsy (ADCME) with complex partial and generalized seizures: A newly recognized epilepsy syndrome with linkage to chromosome 2p11.1-q12.2. Brain 2001, 124, 2459–2475. [Google Scholar] [CrossRef] [Green Version]
- Henden, L.; Freytag, S.; Afawi, Z.; Baldassari, S.; Berkovic, S.F.; Bisulli, F.; Canafoglia, L.; Casari, G.; Crompton, D.E.; Depienne, C.; et al. Identity by descent fine mapping of familial adult myoclonus epilepsy (FAME) to 2p11.2-2q11.2. Hum. Genet. 2016, 135, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Saint-Martin, C.; Bouteiller, D.; Stevanin, G.; Popescu, C.; Charon, C.; Ruberg, M.; Baulac, S.; LeGuern, E.; Labauge, P.; Depienne, C. Refinement of the 2p11.1-q12.2 locus responsible for cortical tremor associated with epilepsy and exclusion of candidate genes. Neurogenetics 2008, 9, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Dolzhenko, E.; van Vugt, J.; Shaw, R.J.; Bekritsky, M.A.; van Blitterswijk, M.; Narzisi, G.; Ajay, S.S.; Rajan, V.; Lajoie, B.R.; Johnson, N.H.; et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017, 27, 1895–1903. [Google Scholar] [CrossRef] [Green Version]
- Tankard, R.M.; Bennett, M.F.; Degorski, P.; Delatycki, M.B.; Lockhart, P.J.; Bahlo, M. Detecting Expansions of Tandem Repeats in Cohorts Sequenced with Short-Read Sequencing Data. Am. J. Hum. Genet. 2018, 103, 858–873. [Google Scholar] [CrossRef] [Green Version]
- Dashnow, H.; Lek, M.; Phipson, B.; Halman, A.; Sadedin, S.; Lonsdale, A.; Davis, M.; Lamont, P.; Clayton, J.S.; Laing, N.G.; et al. STRetch: Detecting and discovering pathogenic short tandem repeat expansions. Genome Biol. 2018, 19, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, K.; Monjo, T.; Hoang, P.H.; Yoshimura, J.; Yurino, H.; Mitsui, J.; Ishiura, H.; Takahashi, Y.; Ichikawa, Y.; Goto, J.; et al. Rapid detection of expanded short tandem repeats in personal genomics using hybrid sequencing. Bioinformatics 2014, 30, 815–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeetong, P.; Ausavarat, S.; Bhidayasiri, R.; Piravej, K.; Pasutharnchat, N.; Desudchit, T.; Chunharas, C.; Loplumlert, J.; Limotai, C.; Suphapeetiporn, K.; et al. A newly identified locus for benign adult familial myoclonic epilepsy on chromosome 3q26.32-3q28. Eur. J. Hum. Genet. 2013, 21, 225–228. [Google Scholar] [CrossRef] [Green Version]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [Green Version]
- Kelkar, Y.D.; Eckert, K.A.; Chiaromonte, F.; Makova, K.D. A matter of life or death: How microsatellites emerge in and vanish from the human genome. Genome Res. 2011, 21, 2038–2048. [Google Scholar] [CrossRef] [Green Version]
- Clark, R.M.; Dalgliesh, G.L.; Endres, D.; Gomez, M.; Taylor, J.; Bidichandani, S.I. Expansion of GAA triplet repeats in the human genome: Unique origin of the FRDA mutation at the center of an Alu. Genomics 2004, 83, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Yang, L. ALUternative Regulation for Gene Expression. Trends Cell Biol. 2017, 27, 480–490. [Google Scholar] [CrossRef] [Green Version]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef]
- Hammond, S.M.; Aartsma-Rus, A.; Alves, S.; Borgos, S.E.; Buijsen, R.A.M.; Collin, R.W.J.; Covello, G.; Denti, M.A.; Desviat, L.R.; Echevarria, L.; et al. Delivery of oligonucleotide-based therapeutics: Challenges and opportunities. EMBO Mol. Med. 2021, 13, e13243. [Google Scholar] [CrossRef]
- Crooke, S.T.; Liang, X.H.; Baker, B.F.; Crooke, R.M. Antisense technology: A review. J. Biol. Chem. 2021, 296, 100416. [Google Scholar] [CrossRef]
- Scoles, D.R.; Minikel, E.V.; Pulst, S.M. Antisense oligonucleotides: A primer. Neurol. Genet. 2019, 5, e323. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.C.; Lobo, D.D.; Martins, I.M.; Lopes, S.M.; Henriques, C.; Duarte, S.P.; Dodart, J.C.; Nobre, R.J.; Pereira de Almeida, L. Antisense oligonucleotide therapeutics in neurodegenerative diseases: The case of polyglutamine disorders. Brain 2020, 143, 407–429. [Google Scholar] [CrossRef]
- Kuijper, E.C.; Bergsma, A.J.; Pijnappel, W.; Aartsma-Rus, A. Opportunities and challenges for antisense oligonucleotide therapies. J. Inherit. Metab. Dis. 2021, 44, 72–87. [Google Scholar] [CrossRef]
- Oliveira, C.; Silveira, I.; Veiga, F.; Ribeiro, A.J. Recent advances in characterization of nonviral vectors for delivery of nucleic acids: Impact on their biological performance. Expert Opin. Drug Deliv. 2015, 12, 27–39. [Google Scholar] [CrossRef]
- Evers, M.M.; Toonen, L.J.; van Roon-Mom, W.M. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv. Drug Deliv. Rev. 2015, 87, 90–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, J.; Kordasiewicz, H.B.; O’Callaghan, B.; Handler, H.P.; Wagener, C.; Duvick, L.; Swayze, E.E.; Rainwater, O.; Hofstra, B.; Benneyworth, M.; et al. Antisense oligonucleotide-mediated ataxin-1 reduction prolongs survival in SCA1 mice and reveals disease-associated transcriptome profiles. JCI Insight 2018, 3, e123193. [Google Scholar] [CrossRef] [PubMed]
- Scoles, D.R.; Meera, P.; Schneider, M.D.; Paul, S.; Dansithong, W.; Figueroa, K.P.; Hung, G.; Rigo, F.; Bennett, C.F.; Otis, T.S.; et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 2017, 544, 362–366. [Google Scholar] [CrossRef]
- McLoughlin, H.S.; Moore, L.R.; Chopra, R.; Komlo, R.; McKenzie, M.; Blumenstein, K.G.; Zhao, H.; Kordasiewicz, H.B.; Shakkottai, V.G.; Paulson, H.L. Oligonucleotide therapy mitigates disease in spinocerebellar ataxia type 3 mice. Ann. Neurol. 2018, 84, 64–77. [Google Scholar] [CrossRef]
- Moore, L.R.; Rajpal, G.; Dillingham, I.T.; Qutob, M.; Blumenstein, K.G.; Gattis, D.; Hung, G.; Kordasiewicz, H.B.; Paulson, H.L.; McLoughlin, H.S. Evaluation of Antisense Oligonucleotides Targeting ATXN3 in SCA3 Mouse Models. Mol. Ther. Nucleic Acids 2017, 7, 200–210. [Google Scholar] [CrossRef] [Green Version]
- Toonen, L.J.; Schmidt, I.; Luijsterburg, M.S.; van Attikum, H.; van Roon-Mom, W.M. Antisense oligonucleotide-mediated exon skipping as a strategy to reduce proteolytic cleavage of ataxin-3. Sci. Rep. 2016, 6, 35200. [Google Scholar] [CrossRef] [Green Version]
- Evers, M.M.; Tran, H.D.; Zalachoras, I.; Pepers, B.A.; Meijer, O.C.; den Dunnen, J.T.; van Ommen, G.J.; Aartsma-Rus, A.; van Roon-Mom, W.M. Ataxin-3 protein modification as a treatment strategy for spinocerebellar ataxia type 3: Removal of the CAG containing exon. Neurobiol. Dis. 2013, 58, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Niu, C.; Prakash, T.P.; Kim, A.; Quach, J.L.; Huryn, L.A.; Yang, Y.; Lopez, E.; Jazayeri, A.; Hung, G.; Sopher, B.L.; et al. Antisense oligonucleotides targeting mutant Ataxin-7 restore visual function in a mouse model of spinocerebellar ataxia type 7. Sci. Transl. Med. 2018, 10, eaap8677. [Google Scholar] [CrossRef] [Green Version]
- Yadava, R.S.; Mandal, M.; Giese, J.M.; Rigo, F.; Bennett, C.F.; Mahadevan, M.S. Modeling muscle regeneration in RNA toxicity mice. Hum. Mol. Genet. 2021, 30, 1111–1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dodart, J.C.; Tran, H.; Berkovitch, S.; Braun, M.; Byrne, M.; Durbin, A.F.; Hu, X.S.; Iwamoto, N.; Jang, H.G.; et al. Variant-selective stereopure oligonucleotides protect against pathologies associated with C9orf72-repeat expansion in preclinical models. Nat. Commun. 2021, 12, 847. [Google Scholar] [CrossRef]
- Jiang, J.; Zhu, Q.; Gendron, T.F.; Saberi, S.; McAlonis-Downes, M.; Seelman, A.; Stauffer, J.E.; Jafar-Nejad, P.; Drenner, K.; Schulte, D.; et al. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron 2016, 90, 535–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toonen, L.J.A.; Rigo, F.; van Attikum, H.; van Roon-Mom, W.M.C. Antisense Oligonucleotide-Mediated Removal of the Polyglutamine Repeat in Spinocerebellar Ataxia Type 3 Mice. Mol. Ther. Nucleic Acids 2017, 8, 232–242. [Google Scholar] [CrossRef] [Green Version]
- Kingwell, K. Double setback for ASO trials in Huntington disease. Nat. Rev. Drug Discov. 2021, 20, 412–413. [Google Scholar] [CrossRef]
- Yang, W.Y.; Gao, R.; Southern, M.; Sarkar, P.S.; Disney, M.D. Design of a bioactive small molecule that targets r(AUUCU) repeats in spinocerebellar ataxia 10. Nat. Commun. 2016, 7, 11647. [Google Scholar] [CrossRef] [PubMed]



| Disease | Type | Gene | Pathogenic Alleles | Non-Pathogenic Alleles | |
|---|---|---|---|---|---|
| Common | Less Common | ||||
| SCA | 27SCA10 | ATXN10 | (ATTCT)800–4500 | (ATTCT)10–16 | (ATTCT)17–32 |
| 34SCA31 | BEAN1 | TAAAA(TAGAA)1–4(TGGAA)n | (TAAAA)8–20 | (TAAAA)exp | |
| (TAAAA)n(TAGAA)n and (TAAAATAGAA)n | |||||
| 35SCA37 | DAB1 | (ATTTT)60–79(ATTTC)31–75(ATTTT)58–90 | (ATTTT)7–30 | (ATTTT)31–400 | |
| CANVAS | 32CANVAS | RFC1 | (AAGGG)400-2000 | (AAAAG)11 | (AAAAG)15–200 |
| (AAAGG)40–1000 | |||||
| 130(ACAGG)~1000 | |||||
| 131(AAAGG)10–25(AAGGG)exp | |||||
| FAME | 36FAME1 | SAMD12 | (ATTTT)exp(ATTTC)exp | (ATTTT)<100 | (ATTTT)>100 |
| [(ATTTT)exp(ATTTC)exp(ATTTT)exp]440–3680 | |||||
| 143(ATTTT)n(ATTTG)n | |||||
| 37FAME2 | STARD7 | (ATTTT)340–390(ATTTC)345–588 | (ATTTT)n | n.d. | |
| 38FAME3 | MARCH6 | [(ATTTT)exp(ATTTC)exp]791–1035 | (ATTTT)9–20 | n.d. | |
| 39FAME4 | YEATS2 | (ATTTT)819(ATTTC)221 | (ATTTT)4–120 | (ATTTT)121–1219 | |
| 36FAME6 | TNRC6A | (ATTTT)22(ATTTC)exp(ATTTT)exp | (ATTTT)<200 bp * | (ATTTT)>200 bp * | |
| 36FAME7 | RAPGEF2 | (ATTTT)exp(ATTTC)exp(ATTTT)n | (ATTTT)<300 bp * | (ATTTT)300–3000 bp * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loureiro, J.R.; Castro, A.F.; Figueiredo, A.S.; Silveira, I. Molecular Mechanisms in Pentanucleotide Repeat Diseases. Cells 2022, 11, 205. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11020205
Loureiro JR, Castro AF, Figueiredo AS, Silveira I. Molecular Mechanisms in Pentanucleotide Repeat Diseases. Cells. 2022; 11(2):205. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11020205
Chicago/Turabian StyleLoureiro, Joana R., Ana F. Castro, Ana S. Figueiredo, and Isabel Silveira. 2022. "Molecular Mechanisms in Pentanucleotide Repeat Diseases" Cells 11, no. 2: 205. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11020205