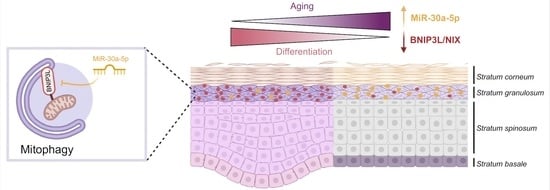
MiR-30a-5p Alters Epidermal Terminal Differentiation during Aging by Regulating BNIP3L/NIX-Dependent Mitophagy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Transfection and Luciferase Assay
2.3. Protein Extraction and Immunoblotting
2.4. Reconstructed Human Epidermis (RHE) Production
2.5. Immunofluorescence
2.6. Total RNA/DNA Isolation and Real-Time Quantitative PCR
2.7. Seahorse Analysis
2.8. Statistical Analysis
3. Results
3.1. BNIP3L Is a New Identified Target of miR-30a-5p
3.2. MiR-30a-5p Abolishes BNIP3L Expression in the Granular Layer of RHE
3.3. BNIP3L Expression Decreases with Chronological Aging in Human Epidermis
3.4. Aging Is Associated with Alterations of Keratinocyte Terminal Differentiation, BNIP3L Expression and Mitochondrial Elimination
3.5. Aged Keratinocytes Display Mitochondrial Metabolic Defects
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pal, S.; Tyler, J.K. Epigenetics and aging. Sci. Adv. 2016, 2, e1600584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saul, D.; Kosinsky, R.L. Epigenetics of Aging and Aging-Associated Diseases. Int. J. Mol. Sci. 2021, 22, E401. [Google Scholar] [CrossRef]
- Chevalier, F.P.; Rorteau, J.; Lamartine, J. MicroRNAs in the Functional Defects of Skin Aging. In Non-Coding RNAs; Tutar, L., Aras, S., Tutar, E., Eds.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef] [Green Version]
- Durante, G.; Comito, F.; Lambertini, M.; Broseghini, E.; Dika, E.; Ferracin, M. Non-coding RNA dysregulation in skin cancers. Essays Biochem. 2021, 65, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Tsoi, L.C.; Gudjonsson, J.E. Dysregulated epigenetic modifications in psoriasis. Exp. Dermatol. 2021, 30, 1156–1166. [Google Scholar] [CrossRef]
- Muther, C.; Jobeili, L.; Garion, M.; Heraud, S.; Thepot, A.; Damour, O.; Lamartine, J. An expression screen for aged-dependent microRNAs identifies miR-30a as a key regulator of aging features in human epidermis. Aging 2017, 9, 2376–2396. [Google Scholar] [CrossRef] [Green Version]
- Monteleon, C.L.; Agnihotri, T.; Dahal, A.; Liu, M.; Rebecca, V.W.; Beatty, G.L.; Amaravadi, R.K.; Ridky, T.W. Lysosomes Support the Degradation, Signaling, and Mitochondrial Metabolism Necessary for Human Epidermal Differentiation. J. Investig. Dermatol. 2018, 138, 1945–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, D.; Cosgarea, I.; Reynolds, N.; Lovat, P.; Armstrong, J. Research Techniques Made Simple: Analysis of Autophagy in the Skin. J. Investig. Dermatol. 2021, 141, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Akinduro, O.; Sully, K.; Patel, A.; Robinson, D.J.; Chikh, A.; McPhail, G.; Braun, K.M.; Philpott, M.P.; Harwood, C.A.; Byrne, C.; et al. Constitutive Autophagy and Nucleophagy during Epidermal Differentiation. J. Investig. Dermatol. 2016, 136, 1460–1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriyama, M.; Moriyama, H.; Uda, J.; Matsuyama, A.; Osawa, M.; Hayakawa, T. BNIP3 plays crucial roles in the differentiation and maintenance of epidermal keratinocytes. J. Investig. Dermatol. 2014, 134, 1627–1635. [Google Scholar] [CrossRef] [Green Version]
- Simpson, C.L.; Tokito, M.K.; Uppala, R.; Sarkar, M.K.; Gudjonsson, J.E.; Holzbaur, E.L.F. NIX initiates mitochondrial fragmentation via DRP1 to drive epidermal differentiation. Cell Rep. 2021, 34, 108689. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, H.; Liu, X.; Li, B.; Chen, Y.; Ren, X.; Liu, C.-G.; Yang, J.-M. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 2009, 5, 816–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pourhanifeh, M.H.; Mahjoubin-Tehran, M.; Karimzadeh, M.R.; Mirzaei, H.R.; Razavi, Z.S.; Sahebkar, A.; Hosseini, N.; Mirzaei, H.; Hamblin, M.R. Autophagy in cancers including brain tumors: Role of MicroRNAs. Cell Commun. Signal. 2020, 18, 88. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Rao, S.; Cao, R.; Xiao, S.; Cui, X.; Ye, L. miR-30a-5p suppresses lung squamous cell carcinoma via ATG5—Mediated autophagy. Aging 2021, 13, 17462–17472. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Deng, X.; Chen, S.; Yang, L.; Ni, J.; Wang, R.; Lin, J.; Bai, M.; Jia, Z.; Huang, S.; et al. MicroRNA-30e targets BNIP3L to protect against aldosterone-induced podocyte apoptosis and mitochondrial dysfunction. Am. J. Physiol.-Ren. Physiol. 2017, 312, F589–F598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, B.; Dai, X.; Li, S.; Qi, G.; Cao, G.; Zhong, Y.; Yin, P.; Yang, X. MiR-30c regulates cisplatin-induced apoptosis of renal tubular epithelial cells by targeting Bnip3L and Hspa5. Cell Death Dis. 2017, 8, e2987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seok, H.; Ham, J.; Jang, E.-S.; Chi, S.W. MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions. Mol. Cells 2016, 39, 375–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahler, E.; Sullivan, W.J.; Cass, A.; Braas, D.; York, A.G.; Bensinger, S.J.; Graeber, T.G.; Christofk, H.R. Doxycycline Alters Metabolism and Proliferation of Human Cell Lines. PLoS ONE 2013, 8, e64561. [Google Scholar] [CrossRef] [PubMed]
- Chinnadurai, G.; Vijayalingam, S.; Gibson, S.B. BNIP3 subfamily BH3-only proteins: Mitochondrial stress sensors in normal and pathological functions. Oncogene 2008, 27, S114–S127. [Google Scholar] [CrossRef] [Green Version]
- Ney, P.A. Mitochondrial autophagy: Origins, significance, and role of BNIP3 and NIX. Biochim Biophys Acta 2015, 1853, 2775–2783. [Google Scholar] [CrossRef] [Green Version]
- Schweers, R.L.; Zhang, J.; Randall, M.S.; Loyd, M.R.; Li, W.; Dorsey, F.C.; Kundu, M.; Opferman, J.T.; Cleveland, J.L.; Miller, J.L.; et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 19500–19505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandoval, H.; Thiagarajan, P.; Dasgupta, S.K.; Schumacher, A.; Prchal, J.T.; Chen, M.; Wang, J. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008, 454, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.A.; McGreal-Estrada, R.; Logan, C.M.; Cvekl, A.; Menko, A.S.; Kantorow, M. BNIP3L/NIX is required for elimination of mitochondria, endoplasmic reticulum and Golgi apparatus during eye lens organelle-free zone formation. Exp. Eye Res. 2018, 174, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Lampert, M.A.; Orogo, A.M.; Najor, R.H.; Hammerling, B.C.; Leon, L.J.; Wang, B.J.; Kim, T.; Sussman, M.A.; Gustafsson, Å.B. BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy 2019, 15, 1182–1198. [Google Scholar] [CrossRef]
- Yazdankhah, M.; Ghosh, S.; Shang, P.; Stepicheva, N.; Hose, S.; Liu, H.; Chamling, X.; Tian, S.; Sullivan, M.L.G.; Calderon, M.J.; et al. BNIP3L-mediated mitophagy is required for mitochondrial remodeling during the differentiation of optic nerve oligodendrocytes. Autophagy 2021, 17, 3140–3159. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Man, M.-Q.; Li, T.; Elias, P.M.; Mauro, T.M. Aging-associated alterations in epidermal function and their clinical significance. Aging 2020, 12, 5551–5565. [Google Scholar] [CrossRef]
- Rinnerthaler, M.; Duschl, J.; Steinbacher, P.; Salzmann, M.; Bischof, J.; Schuller, M.; Wimmer, H.; Peer, T.; Bauer, J.W.; Richter, K. Age-related changes in the composition of the cornified envelope in human skin. Exp. Dermatol. 2013, 22, 329–335. [Google Scholar] [CrossRef]
- Xiaoyun, X.; Chaofei, H.; Weiqi, Z.; Chen, C.; Lixia, L.; Queping, L.; Cong, P.; Shuang, Z.; Juan, S.; Xiang, C. Possible Involvement of F1F0-ATP synthase and Intracellular ATP in Keratinocyte Differentiation in normal skin and skin lesions. Sci. Rep. 2017, 7, 42672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Zhang, M.; Jeong, Y.Y.; Margolis, D.J.; Cai, Q. The role of mitophagy in the regulation of mitochondrial energetic status in neurons. Autophagy 2021, 17, 4182–4201. [Google Scholar] [CrossRef] [PubMed]
- Sgarbi, G.; Matarrese, P.; Pinti, M.; Lanzarini, C.; Ascione, B.; Gibellini, L.; Dika, E.; Patrizi, A.; Tommasino, C.; Capri, M.; et al. Mitochondria hyperfusion and elevated autophagic activity are key mechanisms for cellular bioenergetic preservation in centenarians. Aging 2014, 6, 296–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, S. The Mitochondrial Basis of Aging and Age-Related Disorders. Genes 2017, 8, 398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Dong, Z.; Wang, J.; Tao, Y.; Sun, X.; Yao, X. The existence and function of mitochondrial component in extracellular vesicles. Mitochondrion 2020, 54, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Choong, C.-J.; Okuno, T.; Ikenaka, K.; Baba, K.; Hayakawa, H.; Koike, M.; Yokota, M.; Doi, J.; Kakuda, K.; Takeuchi, T.; et al. Alternative mitochondrial quality control mediated by extracellular release. Autophagy 2020, 17, 2962–2974. [Google Scholar] [CrossRef] [PubMed]
- Lazo, S.; Noren Hooten, N.; Green, J.; Eitan, E.; Mode, N.A.; Liu, Q.-R.; Zonderman, A.B.; Ezike, N.; Mattson, M.P.; Ghosh, P.; et al. Mitochondrial DNA in extracellular vesicles declines with age. Aging Cell 2021, 20, e13283. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Noren Hooten, N.; Evans, M.K. Mitochondria as extracellular vesicle cargo in aging. Aging 2021, 13, 17957–17958. [Google Scholar] [CrossRef]





| Gene | KRT10 | IVL | LOR | TGM1 | FLG | AQP9 | CDSN | KLK7 | BNIP3L | |
|---|---|---|---|---|---|---|---|---|---|---|
| Groups of age | Young | D6 | D6 | D9 | D6 | D9 | D9 | D9 | D9 | D6 |
| 244.7 | 38.7 | 984 | 24.9 | 201.4 | 127.7 | 576.8 | 215.8 | 34.7 | ||
| Adult | D9 | D6 | D6 | D6 | D9 | D6 | D6 | D9 | D9 | |
| 228.5 | 39.8 | 836 | 31.8 | 232.8 | 48.2 | 167.7 | 177.7 | 30.9 | ||
| Aged | D9 | D9 | D9 | D6 | D9 | D9 | D9 | D9 | D9 | |
| 52.4 | 25 | 189.5 | 25.2 | 139.7 | 25.2 | 74.1 | 35.8 | 8.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chevalier, F.P.; Rorteau, J.; Ferraro, S.; Martin, L.S.; Gonzalez-Torres, A.; Berthier, A.; El Kholti, N.; Lamartine, J. MiR-30a-5p Alters Epidermal Terminal Differentiation during Aging by Regulating BNIP3L/NIX-Dependent Mitophagy. Cells 2022, 11, 836. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11050836
Chevalier FP, Rorteau J, Ferraro S, Martin LS, Gonzalez-Torres A, Berthier A, El Kholti N, Lamartine J. MiR-30a-5p Alters Epidermal Terminal Differentiation during Aging by Regulating BNIP3L/NIX-Dependent Mitophagy. Cells. 2022; 11(5):836. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11050836
Chicago/Turabian StyleChevalier, Fabien P., Julie Rorteau, Sandra Ferraro, Lisa S. Martin, Alejandro Gonzalez-Torres, Aurore Berthier, Naima El Kholti, and Jérôme Lamartine. 2022. "MiR-30a-5p Alters Epidermal Terminal Differentiation during Aging by Regulating BNIP3L/NIX-Dependent Mitophagy" Cells 11, no. 5: 836. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11050836