Alterations of Mitochondrial Biology in the Oral Mucosa of Chilean Children with Autism Spectrum Disorder (ASD)
Abstract
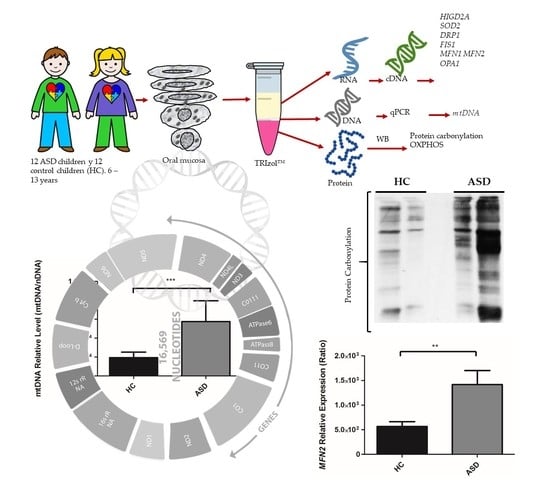
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Subjects
2.3. Oral Sample Preparation
2.4. Isolation of RNA, DNA, and Proteins
2.5. Reverse Transcriptase and Quantitative Real-Time PCR (qRT-PCR)
2.6. qPCR of mtDNA Levels
2.7. Tetra-Primer Amplification-Refractory Mutation System (ARMS)-PCR
2.8. Western Blot
2.9. Protein Oxidation Detection
2.10. Statistical Analysis
3. Results
3.1. Subjects Clinical Data and Demographic Information
3.2. Oral Mucosa mtDNA Levels in Chilean Children with ASD
3.3. Oral Mucosa Total Protein Oxidation Levels and Western Blotting Analysis of Respiratory Complexes in Chilean Children with ASD
3.4. The Presence of Ala16val-SOD2 Polymorphism in Chilean Children with ASD
3.5. Expression of the Genes Encoding for HIGD2A, SOD2, DRP1, FIS1, MFN1, MFN2 and OPA1 from the Oral Mucosa of Chilean Children with ASD
3.6. Prediction of the Association of the Analyzed Genes with the Genetic Bases of ASD
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wing, L.; Gould, J. Severe impairments of social interaction and associated abnormalities in children: Epidemiology and classification. J. Autism Dev. Disord. 1979, 9, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Wing, L. The continuum of autistic characteristics. In Diagnosis and Assessment in Autism; Springer: Boston, MA, USA, 1988; pp. 91–110. [Google Scholar]
- Borden, M.C.; Ollendick, T.H. An examination of the validity of social subtypes in autism. J. Autism Dev. Disord. 1994, 24, 23–37. [Google Scholar] [CrossRef]
- Eisenmajer, R.; Prior, M.; Leekam, S.; Wing, L.; Gould, J.; Welham, M.; Ong, B. Comparison of clinical symptoms in autism and Asperger’s disorder. J. Am. Acad. Child Adolesc. Psychiatry 1996, 35, 1523–1531. [Google Scholar] [CrossRef]
- Baird, G.; Simonoff, E.; Pickles, A.; Chandler, S.; Loucas, T.; Meldrum, D.; Charman, T. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: The Special Needs and Autism Project (SNAP). Lancet 2006, 368, 210–215. [Google Scholar] [CrossRef]
- Gabriels, R.L.; Agnew, J.A.; Miller, L.J.; Gralla, J.; Pan, Z.; Goldson, E.; Ledbetter, J.C.; Dinkins, J.P.; Hooks, E. Is there a relationship between restricted, repetitive, stereotyped behaviors and interests and abnormal sensory response in children with autism spectrum disorders? Res. Autism Spectr. Disord. 2008, 2, 660–670. [Google Scholar] [CrossRef]
- Weintraub, K. Autism counts. Nature 2011, 479, 22. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.L.; Bilder, D.A.; Zahorodny, W.; Pettygrove, S.; Durkin, M.S.; Fitzgerald, R.T.; Rice, C.; Kurzius-Spencer, M.; Baio, J.; Yeargin-Allsopp, M. Prevalence and characteristics of autism spectrum disorder among 4-year-old children in the autism and developmental disabilities monitoring network. J. Dev. Behav. Pediatr. 2016, 37, 1–8. [Google Scholar] [CrossRef]
- Rose, S.; Niyazov, D.M.; Rossignol, D.A.; Goldenthal, M.; Kahler, S.G.; Frye, R.E. Clinical and Molecular Characteristics of Mitochondrial Dysfunction in Autism Spectrum Disorder. Mol. Diagn. Ther. 2018, 22, 571–593. [Google Scholar] [CrossRef]
- Dawson, G. Early behavioral intervention, brain plasticity, and the prevention of autism spectrum disorder. Dev. Psychopathol. 2008, 20, 775–803. [Google Scholar] [CrossRef]
- Warren, Z.; McPheeters, M.L.; Sathe, N.; Foss-Feig, J.H.; Glasser, A.; Veenstra-VanderWeele, J. A systematic review of early intensive intervention for autism spectrum disorders. Pediatrics 2011, 127, e1303–e1311. [Google Scholar] [CrossRef]
- Chauhan, A.; Chauhan, V. Oxidative stress in autism. Pathophysiology 2006, 13, 171–181. [Google Scholar] [CrossRef]
- László, A.; Horváth, E.; Eck, E.; Fekete, M. Serum serotonin, lactate and pyruvate levels in infantile autistic children. Clin. Chim. Acta 1994, 229, 205–207. [Google Scholar] [CrossRef]
- Dhillon, S.; Hellings, J.A.; Butler, M.G. Genetics and mitochondrial abnormalities in autism spectrum disorders: A review. Curr. Genom. 2011, 12, 322–332. [Google Scholar] [CrossRef]
- Rossignol, D.A.; Frye, R.E. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol. Psychiatry 2012, 17, 290–314. [Google Scholar] [CrossRef]
- Gu, F.; Chauhan, V.; Kaur, K.; Brown, W.T.; LaFauci, G.; Wegiel, J.; Chauhan, A. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl. Psychiatry 2013, 3, e299. [Google Scholar] [CrossRef]
- MacFabe, D. Autism: Metabolism, Mitochondria, and the Microbiome. Glob. Adv. Health Med. 2013, 2, 52–66. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Rios, P.G.; Kuo, S.-H.; Akman, H.O.; Rosoklija, G.; Tanji, K.; Dwork, A.; Schon, E.A.; DiMauro, S.; Goldman, J. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol. Dis. 2013, 54, 349–361. [Google Scholar] [CrossRef] [Green Version]
- Sutton, A.; Imbert, A.; Igoudjil, A.; Descatoire, V.; Cazanave, S.; Pessayre, D.; Degoul, F. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharmacogenet. Genom. 2005, 15, 311–319. [Google Scholar] [CrossRef]
- Esparham, A.E.; Smith, T.; Belmont, J.M.; Haden, M.; Wagner, L.E.; Evans, R.G.; Drisko, J.A. Nutritional and Metabolic Biomarkers in Autism Spectrum Disorders: An Exploratory Study. Integr. Med. A Clin. J. 2015, 14, 40–53. [Google Scholar]
- Giulivi, C.; Zhang, Y.-F.; Omanska-Klusek, A.; Ross-Inta, C.; Wong, S.; Hertz-Picciotto, I.; Tassone, F.; Pessah, I.N. Mitochondrial dysfunction in autism. JAMA 2010, 304, 2389–2396. [Google Scholar] [CrossRef]
- Chen, S.; Li, Z.; He, Y.; Zhang, F.; Li, H.; Liao, Y.; Wei, Z.; Wan, G.; Xiang, X.; Hu, M.; et al. Elevated mitochondrial DNA copy number in peripheral blood cells is associated with childhood autism. BMC Psychiatry 2015, 15, 50. [Google Scholar] [CrossRef]
- Hadjixenofontos, A.; Schmidt, M.A.; Whitehead, P.L.; Konidari, I.; Hedges, D.J.; Wright, H.H.; Abramson, R.K.; Menon, R.; Williams, S.M.; Cuccaro, M.L. Evaluating mitochondrial DNA variation in autism spectrum disorders. Ann. Hum. Genet. 2013, 77, 9–21. [Google Scholar] [CrossRef]
- Jones, K.B.; Klein, O.D. Oral epithelial stem cells in tissue maintenance and disease: The first steps in a long journey. Int. J. Oral Sci. 2013, 5, 121–129. [Google Scholar] [CrossRef]
- Goldenthal, M.J.; Kuruvilla, T.; Damle, S.; Salganicoff, L.; Sheth, S.; Shah, N.; Marks, H.; Khurana, D.; Valencia, I.; Legido, A. Non-invasive evaluation of buccal respiratory chain enzyme dysfunction in mitochondrial disease: Comparison with studies in muscle biopsy. Mol. Genet. Metab. 2012, 105, 457–462. [Google Scholar] [CrossRef]
- Goldenthal, M.J.; Damle, S.; Sheth, S.; Shah, N.; Melvin, J.; Jethva, R.; Hardison, H.; Marks, H.; Legido, A. Mitochondrial enzyme dysfunction in autism spectrum disorders; a novel biomarker revealed from buccal swab analysis. Biomark. Med. 2015, 9, 957–965. [Google Scholar] [CrossRef]
- Burger, B.J.; Rose, S.; Bennuri, S.C.; Gill, P.S.; Tippett, M.L.; Delhey, L.; Melnyk, S.; Frye, R.E. Autistic Siblings with Novel Mutations in Two Different Genes: Insight for Genetic Workups of Autistic Siblings and Connection to Mitochondrial Dysfunction. Front. Pediatr. 2017, 5, 219. [Google Scholar] [CrossRef]
- Frye, R.E.; DeLaTorre, R.; Taylor, H.; Slattery, J.; Melnyk, S.; Chowdhury, N.; James, S.J. Redox metabolism abnormalities in autistic children associated with mitochondrial disease. Transl. Psychiatry 2013, 3, e273. [Google Scholar] [CrossRef]
- Delhey, L.M.; Nur Kilinc, E.; Yin, L.; Slattery, J.C.; Tippett, M.L.; Rose, S.; Bennuri, S.C.; Kahler, S.G.; Damle, S.; Legido, A.; et al. The Effect of Mitochondrial Supplements on Mitochondrial Activity in Children with Autism Spectrum Disorder. J. Clin. Med. 2017, 6, 18. [Google Scholar] [CrossRef]
- Rose, S.; Bennuri, S.C.; Davis, J.E.; Wynne, R.; Slattery, J.C.; Tippett, M.; Delhey, L.; Melnyk, S.; Kahler, S.G.; MacFabe, D.F.; et al. Butyrate enhances mitochondrial function during oxidative stress in cell lines from boys with autism. Transl. Psychiatry 2018, 8, 42. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Taylor, E.B.; Dephoure, N.; Heo, J.-M.; Tonhato, A.; Papandreou, I.; Nath, N.; Denko, N.C.; Gygi, S.P.; Rutter, J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012, 15, 348–360. [Google Scholar] [CrossRef] [Green Version]
- Acín-Pérez, R.; Fernández-Silva, P.; Peleato, M.L.; Pérez-Martos, A.; Enriquez, J.A. Respiratory Active Mitochondrial Supercomplexes. Mol. Cell 2008, 32, 529–539. [Google Scholar] [CrossRef]
- Salazar, C.; Elorza, A.A.; Cofre, G.; Ruiz-Hincapie, P.; Shirihai, O.; Ruiz, L.M. The OXPHOS supercomplex assembly factor HIG2A responds to changes in energetic metabolism and cell cycle. J. Cell. Physiol. 2019, 0, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rooney, J.P.; Ryde, I.T.; Sanders, L.H.; Howlett, E.H.; Colton, M.D.; Germ, K.E.; Mayer, G.D.; Greenamyre, J.T.; Meyer, J.N. PCR based determination of mitochondrial DNA copy number in multiple species. Mitochondrial Regul. Methods Protoc. 2015, 23–38. [Google Scholar]
- Venegas, V.; Halberg, M.C. Measurement of mitochondrial DNA copy number. Mitochondrial Dis. Biochem. Mol. Anal. 2012, 327–335. [Google Scholar]
- Barbisan, F.; de Rosso Motta, J.; Trott, A.; Azzolin, V.; Dornelles, E.B.; Marcon, M.; Algarve, T.D.; Duarte, M.M.M.F.; Mostardeiro, C.P.; Unfer, T.C. Methotrexate-related response on human peripheral blood mononuclear cells may be modulated by the Ala16Val-SOD2 gene polymorphism. PLoS ONE 2014, 9, e107299. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.M.; Jensen, E.L.; Rossel, Y.; Puas, G.I.; Gonzalez-Ibanez, A.M.; Bustos, R.I.; Ferrick, D.A.; Elorza, A.A. Non-cytotoxic copper overload boosts mitochondrial energy metabolism to modulate cell proliferation and differentiation in the human erythroleukemic cell line K562. Mitochondrion 2016, 29, 18–30. [Google Scholar] [CrossRef]
- Korhonen, J.A.; Pham, X.H.; Pellegrini, M.; Falkenberg, M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004, 23, 2423–2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suomalainen, A.; Battersby, B.J. Mitochondrial diseases: The contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol. 2017, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Valenti, L.; Conte, D.; Piperno, A.; Dongiovanni, P.; Fracanzani, A.; Fraquelli, M.; Vergani, A.; Gianni, C.; Carmagnola, L.; Fargion, S. The mitochondrial superoxide dismutase A16V polymorphism in the cardiomyopathy associated with hereditary haemochromatosis. J. Med. Genet. 2004, 41, 946–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landi, S.; Gemignani, F.; Neri, M.; Barale, R.; Bonassi, S.; Bottari, F.; Canessa, P.A.; Canzian, F.; Ceppi, M.; Filiberti, R. Polymorphisms of glutathione-S-transferase M1 and manganese superoxide dismutase are associated with the risk of malignant pleural mesothelioma. Int. J. Cancer 2007, 120, 2739–2743. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.-Y.; Neuhouser, M.L.; Barnett, M.J.; Hong, C.-C.; Kristal, A.R.; Thornquist, M.D.; King, I.B.; Goodman, G.E.; Ambrosone, C.B. Iron intake, oxidative stress-related genes (MnSOD and MPO) and prostate cancer risk in CARET cohort. Carcinogenesis 2008, 29, 964–970. [Google Scholar] [CrossRef] [Green Version]
- Burri, R.J.; Stock, R.G.; Cesaretti, J.A.; Atencio, D.P.; Peters, S.; Peters, C.A.; Fan, G.; Stone, N.N.; Ostrer, H.; Rosenstein, B.S. Association of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancer. Radiat. Res. 2008, 170, 49–59. [Google Scholar] [CrossRef]
- Bastaki, M.; Huen, K.; Manzanillo, P.; Chande, N.; Chen, C.; Balmes, J.R.; Tager, I.B.; Holland, N. Genotype–activity relationship for Mn-superoxide dismutase, glutathione peroxidase 1 and catalase in humans. Pharmacogenet. Genom. 2006, 16, 279–286. [Google Scholar] [CrossRef]
- Rajaraman, P.; Hutchinson, A.; Rothman, N.; Black, P.M.; Fine, H.A.; Loeffler, J.S.; Selker, R.G.; Shapiro, W.R.; Linet, M.S.; Inskip, P.D. Oxidative response gene polymorphisms and risk of adult brain tumors. Neuro-oncology 2008, 10, 709–715. [Google Scholar] [CrossRef] [Green Version]
- Clair, D.K.S.; Holland, J.C. Complementary DNA encoding human colon cancer manganese superoxide dismutase and the expression of its gene in human cells. Cancer Res. 1991, 51, 939–943. [Google Scholar]
- McCord, J.M.; Fridovich, I. Superoxide dismutase an enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar]
- An, H.-J.; Shin, H.; Jo, S.-G.; Kim, Y.J.; Lee, J.-O.; Paik, S.-G.; Lee, H. The survival effect of mitochondrial Higd-1a is associated with suppression of cytochrome C release and prevention of caspase activation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2011, 1813, 2088–2098. [Google Scholar] [CrossRef] [Green Version]
- Vukotic, M.; Oeljeklaus, S.; Wiese, S.; Vögtle, F.N.; Meisinger, C.; Meyer, H.E.; Zieseniss, A.; Katschinski, D.M.; Jans, D.C.; Jakobs, S.; Warscheid, B.; Rehling, P.; Deckers, M. Rcf1 Mediates Cytochrome Oxidase Assembly and Respirasome Formation, Revealing Heterogeneity of the Enzyme Complex. Cell Metab. 2012, 15, 336–347. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, A.; Zhang, R.; Yao, V.; Theesfeld, C.L.; Wong, A.K.; Tadych, A.; Volfovsky, N.; Packer, A.; Lash, A.; Troyanskaya, O.G. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat. Neurosci. 2016, 19, 1454. [Google Scholar] [CrossRef]
- Hartmann, K.; Kozikowski, C.T.; Urbano, M.R.; Williams, T.V.; Ba, C.L.-T.; Peterkin, A. Autism spectrum disorder in Latin American families: Experiences in Chile. Fam. Syst. Health 2018, 36, 169–174. [Google Scholar] [CrossRef]
- Parisi, M.A.; Clayton, D.A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 1991, 252, 965. [Google Scholar] [CrossRef]
- Larsson, N.-G.; Wang, J.; Wilhelmsson, H.; Oldfors, A.; Rustin, P.; Lewandoski, M.; Barsh, G.S.; Clayton, D.A. Mitochondrial transcription factor A is necessary for mtDNA maintance and embryogenesis in mice. Nature Genet. 1998, 18, 231. [Google Scholar] [CrossRef]
- Zhang, Y.; Qu, Y.; Gao, K.; Yang, Q.; Shi, B.; Hou, P.; Ji, M. High copy number of mitochondrial DNA (mtDNA) predicts good prognosis in glioma patients. Am. J. Cancer Res. 2015, 5, 1207–1216. [Google Scholar]
- Ikeuchi, M.; Matsusaka, H.; Kang, D.; Matsushima, S.; Ide, T.; Kubota, T.; Fujiwara, T.; Hamasaki, N.; Takeshita, A.; Sunagawa, K.; et al. Overexpression of Mitochondrial Transcription Factor A Ameliorates Mitochondrial Deficiencies and Cardiac Failure After Myocardial Infarction. Circulation 2005, 112, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Yoshida, M.; Yamato, M.; Ide, T.; Wu, Z.; Ochi-Shindou, M.; Kanki, T.; Kang, D.; Sunagawa, K.; Tsutsui, H.; et al. Reverse of Age-Dependent Memory Impairment and Mitochondrial DNA Damage in Microglia by an Overexpression of Human Mitochondrial Transcription Factor A in Mice. J. Neurosci. 2008, 28, 8624. [Google Scholar] [CrossRef]
- Ylikallio, E.; Tyynismaa, H.; Tsutsui, H.; Ide, T.; Suomalainen, A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 2010, 19, 2695–2705. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, D.A.; Frye, R.E. Evidence linking oxidative stress, mitochondrial dysfunction, and inflammation in the brain of individuals with autism. Front. Physiol. 2014, 5, 150. [Google Scholar] [CrossRef]
- Anitha, A.; Nakamura, K.; Thanseem, I.; Yamada, K.; Iwayama, Y.; Toyota, T.; Matsuzaki, H.; Miyachi, T.; Yamada, S.; Tsujii, M.; et al. Brain region-specific altered expression and association of mitochondria-related genes in autism. Mol. Autism 2012, 3, 12. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [Green Version]






| Code | Gender | Diagnosis (Age Years) | DSM-IV | DSM-5 (Level) | Education 1 | Therapy 1 | Sample (Age Years) |
|---|---|---|---|---|---|---|---|
| ASD1 | Male | ASD (3) | Severe | III *** | Pre-school | B 2, C 3, S 4 | 9 |
| ASD2 | Male | ASD (4) | Moderate | II ** | Pre-school | B, C, S | 9 |
| ASD3 | Male | ASD (3) | Moderate | II | Pre-school | B, C, S | 8 |
| ASD4 | Male | ASD (4) | Moderate | II | Pre-school | B, C, S | 8 |
| ASD5 | Male | ASD (3) | Moderate | II | Primary school | B, C, S | 12 |
| ASD6 | Male | ASD (5) | Moderate | II | Primary school | B, C, S | 11 |
| ASD7 | Male | ASD (1) | Moderate | II | Pre-school | B, C, S | 7 |
| ASD8 | Male | ASD (2) | Mild | I * | Pre-school | B, C, S | 7 |
| ASD9 | Female | ASD (6) | Moderate | II | Primary school | B, C, S | 11 |
| ASD10 | Male | ASD (1) | Moderate | II | Primary school | B, C, S | 9 |
| ASD11 | Male | ASD (2) | Severe | III | Primary school | B, C, S | 12 |
| ASD12 | Male | ASD (2) | Severe | III | Primary school | B, C, S | 12 |
| Gene | Gene Description | Chr 1 | Edge Score | Rank 2 |
|---|---|---|---|---|
| NFYC | Nuclear transcription factor Y, gamma | 1p34.2 | 0.441 | 2752 |
| OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | 7p13 | 0.401 | 3680 |
| ACO2 | Aconitase 2, mitochondrial | 22q13.2 | 0.395 | 7261 |
| EIF4G1 | Eukaryotic translation initiation factor 4 gamma, 1 | 3q27.1 | 0.352 | 3374 |
| ILF3 | Interleukin enhancer binding factor 3, 90 kDa | 19p13.2 | 0.351 | 2897 |
| CLPTM1 | Cleft lip and palate associated transmembrane protein 1 | 19q13.32 | 0.329 | 355 |
| VAMP3 | Vesicle-associated membrane protein 3 | 1p36.23 | 0.328 | 24,000 |
| ARF1 | ADP-ribosylation factor 1 | 1q42.13 | 0.326 | 1225 |
| UBE4B | Ubiquitination factor E4B | 1p36.22 | 0.322 | 2086 |
| UPF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | 19p13.11 | 0.318 | 2170 |
| CAPZB | Capping protein (actin filament) muscle Z-line, beta | 1p36.13 | 0.316 | 2179 |
| NUDC | NudC nuclear distribution protein | 1p36.11 | 0.313 | 23,418 |
| XPO7 | Exportin 7 | 8p21.3 | 0.303 | 1922 |
| NUP98 | Nucleoporin 98 kDa | 11p15.4 | 0.295 | 7869 |
| ATP6V0B | ATPase, H+ transporting, lysosomal 21 kDa, V0 subunit b | 1p34.1 | 0.294 | 277 |
| DLAT | Dihydrolipoamide S-acetyltransferase | 11q23.1 | 0.289 | 24,135 |
| BSG | Basigin (Ok blood group) | 19p13.3 | 0.289 | 1821 |
| VARS | Valyl-tRNA synthetase | 6p21.33 | 0.282 | 21,435 |
| NSFL1C | NSFL1 (p97) cofactor (p47) | 20p13 | 0.281 | 2031 |
| PRUNE | Prune exopolyphosphatase | 1q21.3 | 0.28 | 10,264 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrasco, M.; Salazar, C.; Tiznado, W.; Ruiz, L.M. Alterations of Mitochondrial Biology in the Oral Mucosa of Chilean Children with Autism Spectrum Disorder (ASD). Cells 2019, 8, 367. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8040367
Carrasco M, Salazar C, Tiznado W, Ruiz LM. Alterations of Mitochondrial Biology in the Oral Mucosa of Chilean Children with Autism Spectrum Disorder (ASD). Cells. 2019; 8(4):367. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8040367
Chicago/Turabian StyleCarrasco, Manuel, Celia Salazar, William Tiznado, and Lina María Ruiz. 2019. "Alterations of Mitochondrial Biology in the Oral Mucosa of Chilean Children with Autism Spectrum Disorder (ASD)" Cells 8, no. 4: 367. https://0-doi-org.brum.beds.ac.uk/10.3390/cells8040367