Fructans Are Differentially Distributed in Root Tissues of Asparagus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Tissue Preparation
2.2. Mass Spectrometry Imaging
2.3. Oligosaccharide Extraction and Profiling
2.4. Proteome Profiling
3. Results
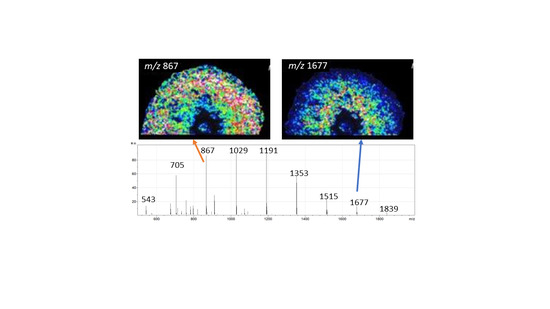
3.1. Mass Spectrometry Imaging Revealed Differential Tissue Abundance Pattern for Fructans
3.2. HPAEC–PAD Analysis Validated Differential Fructan Profiles in the Various Root Tissues
3.3. Proteome Analysis Revealed a Tissue-Specificity in Sucrose Metabolism and Fructan Biosynthesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jiménez-Sánchez, C.J.; Lozano-Sánchez, C.; Rodríguez-Pérez, A.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Comprehensive, untargeted, and qualitative RP-HPLC-ESI-QTOF/MS2 metabolite profiling of green asparagus (Asparagus officinalis). J. Food Comp. Anal. 2016, 46, 78–87. [Google Scholar]
- Zhang, H.; Birch, J.; Pei, J.; Ma, Z.F.; Bekhi, A.E.-D. Phytochemical compounds and biological activity in asparagus roots: A review. Int. J. Food Sci. Tech. 2019, 54, 966–977. [Google Scholar] [CrossRef]
- Shiomi, N.; Yamada, J.; Izawa, M. Isolation and identification of fructo-oligosaccharides in roots of asparagus (Asparagus officinalis L). Agric. Biol. Chem. 1976, 40, 567–575. [Google Scholar] [CrossRef]
- Shiomi, N.; Yamada, J.; Izawa, M. Synthesis of several fructo-oligosaccharides by asparagus fructosyltransferases. Agric. Biol. Chem. 1979, 43, 2233–2244. [Google Scholar]
- Matros, A.; Peukert, M.; Lahnstein, J.; Seiffert, U.; Burton, R. Determination of fructans in plants: Current analytical means for extraction, detection, and quantification. Annu. Plant Rev. Online 2019, 117–156. [Google Scholar]
- Van den Ende, W. Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 2013, 4, 11. [Google Scholar]
- Shiomi, N. Structure of fructopolysaccharide (asparagosin) from roots of asparagus (Asparagus officinalis L.). New Phytol. 1993, 123, 263–270. [Google Scholar] [CrossRef]
- Shiomi, N. Properties of fructosyltransferases involved in the synthesis of fructan in liliaceous plants. J. Plant Physiol. 1989, 134, 151–155. [Google Scholar] [CrossRef]
- Ueno, K.; Sonoda, T.; Yoshida, M.; Shiomi, N.; Onodera, S. Purification, characterization, and functional analysis of a novel 6G&1-FEH mainly hydrolyzing neokestose from asparagus. J. Exp. Bot. 2018, 69, 4295–4308. [Google Scholar]
- Ueno, K.; Onodera, S.; Kawakami, A.; Yoshida, M.; Shiomi, N. Molecular characterization and expression of a cDNA encoding fructan: Fructan 6(G)-fructosyltransferase from asparagus (Asparagus officinalis). New Phytol. 2005, 165, 813–824. [Google Scholar] [CrossRef]
- Amiard, V.; Morvan-Bertrand, A.; Billard, J.-P.; Huault, C.; Keller, F.; Prud’homme, M.-P. Fructans, but not the sucrosyl-galactosides, raffinose and loliose, are affected by drought stress in perennial ryegrass. Plant Physiol. 2003, 132, 2218–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salinas, C.; Handford, M.; Pauly, M.; Dupree, P.; Cardemil, L. Structural modifications of fructans in Aloe barbadensis Miller (Aloe vera) grown under water stress. PLoS ONE 2016, 11, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abeynayake, S.W.; Etzerodt, T.P.; Jonaviciene, K.; Byrne, S.; Asp, T.; Boelt, B. Fructan metabolism and changes in fructan composition during cold acclimation in perennial ryegrass. Front. Plant Sci. 2015, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bie, X.M.; Wang, K.; She, M.Y.; Du, L.P.; Zhang, S.X.; Li, J.R.; Gao, X.; Lin, Z.S.; Ye, X.G. Combinational transformation of three wheat genes encoding fructan biosynthesis enzymes confers increased fructan content and tolerance to abiotic stresses in tobacco. Plant Cell Rep. 2012, 31, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Livingston, D.P.; Hincha, D.K.; Heyer, A.G. Fructan and its relationship to abiotic stress tolerance in plants. Cell. Mol. Life Sci. 2009, 66, 2007–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valluru, R.; Van den Ende, W. Plant fructans in stress environments: Emerging concepts and future prospects. J. Exp. Bot. 2008, 59, 2905–2916. [Google Scholar] [CrossRef] [Green Version]
- Peukert, M.; Thiel, J.; Peshev, D.; Weschke, W.; Van den Ende, W.; Mock, H.-P.; Matros, A. Spatio-temporal dynamics of fructan metabolism in developing barley grains. Plant Cell 2014, 26, 3728–3744. [Google Scholar] [CrossRef] [Green Version]
- Vergauwen, R.; Van den Ende, W.; Van Laere, A. The role of fructan in flowering of Campanula rapunculoides. J. Exp. Bot. 2000, 51, 1261–1266. [Google Scholar] [CrossRef] [Green Version]
- Shiomi, N.; Benkeblia, N.; Onodera, S.; Kawazoe, N. Fructooligosaccharides changes during maturation in inflorescences and seeds of onion (Allium cepa L. ‘W202’). Can. J. Plant Sci. 2006, 86, 269–278. [Google Scholar] [CrossRef]
- Bieleski, R.L. Fructan hydrolysis drives petal expansion in the ephemeral daylily flower. Plant Physiol. 1993, 103, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Solhaug, K.A.; Aares, E. Remobilization of fructans in Phippsia algida during rapid inflorescence development. Physiol. Plant. 1994, 91, 219–225. [Google Scholar] [CrossRef]
- Portes, M.T.; Carvalho, M.A.M. Spatial distribution of fructans and fructan metabolizing enzymes in rhizophores of Vernonia herbacea (Vell.) Rusby (Asteraceae) in different developmental phases. Plant Sci. 2006, 170, 624–633. [Google Scholar] [CrossRef]
- Rigui, A.P.; Gaspar, M.; Oliveira, V.F.; Purgatto, E.; de Carvalho, M.A.M. Endogenous hormone concentrations correlate with fructan metabolism throughout the phenological cycle in Chrysolaena obovata. Ann. Bot. 2015, 115, 1163–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Ende, W.; Michiels, A.; Van Wonterghem, D.; Vergauwen, R.; Van Laere, A. Cloning, developmental, and tissue-specific expression of sucrose: Sucrose 1-fructosyl transferase from Taraxacum officinale. Fructan localization in roots. Plant Physiol. 2000, 123, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Vilhalva, D.A.A.; Cortelazzo, A.L.; Carvalho, A.; Figueiredo-Ribeiro, R.D.L. Histochemistry and ultrastructure of Campuloclinium chlorolepis (Asteraceae) tuberous roots accumulating fructan: Evidences of functions other than reserve carbohydrate. Aust. J. Bot. 2011, 59, 46–52. [Google Scholar] [CrossRef]
- Joaquim, E.O.; Hayashi, A.H.; Torres, L.M.B.; Figueiredo-Ribeiro, R.C.L.; Shiomi, N.; de Sousa, F.S.; Lago, J.H.G.; Carvalho, M.A.M. Chemical structure and localization of levan, the predominant fructan type in underground systems of Gomphrena marginata (Amaranthaceae). Front. Plant Sci. 2018, 9, 10. [Google Scholar] [CrossRef]
- Qin, L.; Zhang, Y.W.; Liu, Y.Q.; He, H.X.; Han, M.M.; Li, Y.Y.; Zeng, M.M.; Wang, X.D. Recent advances in matrix-assisted laser desorption/ionisation mass spectrometry imaging (MALDI-MSI) for in situ analysis of endogenous molecules in plants. Phytochem. Anal. 2018, 29, 351–364. [Google Scholar] [CrossRef]
- Velickovic, D.; Ropartz, D.; Guillon, F.; Saulnier, L.; Rogniaux, H. New insights into the structural and spatial variability of cell-wall polysaccharides during wheat grain development, as revealed through MALDI mass spectrometry imaging. J. Exp. Bot. 2014, 65, 2079–2091. [Google Scholar] [CrossRef] [Green Version]
- Peukert, M.; Lim, W.L.; Seiffert, U.; Matros, A. Mass spectrometry imaging of metabolites in barley grain tissues. Curr. Protoc. Plant Biol. 2016, 1, 574–591. [Google Scholar] [CrossRef]
- Nourbakhsh-Rey, M.; Libault, M. Decipher the molecular response of plant single cell types to environmental stresses. Biomed. Res. Int. 2016, 4182071. [Google Scholar] [CrossRef] [Green Version]
- Szymanski, J.; Levin, Y.; Savidor, A.; Breitel, D.; Chappell-Maor, L.; Heinig, U.; Topfer, N.; Aharoni, A. Label-free deep shotgun proteomics reveals protein dynamics during tomato fruit tissues development. Plant J. 2017, 90, 396–417. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, H.; Bhatti, S.; Zhou, S.; Yang, Y.; Fish, T.; Thannhauser, T.W. Development of a laser capture microscope-based single-cell-type proteomics tool for studying proteomes of individual cell layers of plant roots. Hortic. Res. 2016, 3, 16026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djalali Farahani-Kofoet, R.; Witzel, K.; Gräfe, J.; Grosch, R.; Zrenner, R. Species-specific impact of Fusarium infection on the root and shoot characteristics of asparagus. Pathogens 2020, 9, 509. [Google Scholar] [CrossRef] [PubMed]
- Verspreet, J.; Pollet, A.; Cuyvers, S.; Vergauwen, R.; van den Ende, W.; Delcour, J.A.; Courtin, C.M. A simple and accurate method for determining wheat grain fructan content and average degree of polymerization. J. Agric. Food Chem. 2012, 60, 2102–2107. [Google Scholar] [CrossRef] [PubMed]
- Matros, A.; Houston, K.; Tucker, M.R.; Schreiber, M.; Berger, B.; Aubert, M.K.; Wilkinson, L.G.; Witzel, K.; Waugh, R.; Seiffert, U.; et al. GWAS reveals the genetic complexity of fructan accumulation patterns in barley grain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yamamori, A.; Okada, H.; Kawazoe, N.; Ueno, K.; Onodera, S.; Shiomi, N. Structure of fructan prepared from onion bulbs (Allium cepa L.). J. Appl. Glycosci. 2015, 62, 95–99. [Google Scholar] [CrossRef] [Green Version]
- Verspreet, J.; Dornez, E.; Delcour, J.A.; Harrison, S.J.; Courtin, C.M. Purification of wheat grain fructans from wheat bran. J. Cereal Sci. 2015, 65, 57–59. [Google Scholar] [CrossRef]
- Verspreet, J.; Hemdane, S.; Dornez, E.; Cuyvers, S.; Pollet, A.; Delcour, J.A.; Courtin, C.M. Analysis of storage and structural carbohydrates in developing wheat (Triticum aestivum L.) grains using quantitative analysis and microscopy. J. Agric. Food Chem. 2013, 61, 9251–9259. [Google Scholar] [CrossRef]
- Kaspar, S.; Weier, D.; Weschke, W.; Mock, H.P.; Matros, A. Protein analysis of laser capture micro-dissected tissues revealed cell-type specific biological functions in developing barley grains. Anal. Bioanal. Chem. 2010, 398, 2883–2893. [Google Scholar] [CrossRef]
- Witzel, K.; Risha, M.A.; Albers, P.; Börnke, F.; Hanschen, F.S. Identification and characterization of three epithiospecifier protein isoforms in Brassica oleracea. Front. Plant Sci. 2019, 10, 1552. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Shiomi, N. Content of carbohydrate and activities of fructosyltransferase and invertase in asparagus roots during the fructo-oligosaccharide-accumulating and fructo-polysaccharide-accumulating season. New Phytol. 1992, 122, 421–432. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Hashimoto, K.; Toyooka, K.; Saito, K. Keeping the shape of plant tissue for visualizing metabolite features in segmentation and correlation analysis of imaging mass spectrometry in Asparagus officinalis. Metabolomics 2019, 15, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi, K.; Enomoto, Y.; Fukusaki, E.; Shimma, S. Visualization of asparaptine in asparagus (Asparagus officinalis) using MALDI-IMS. Anal. Sci. 2018, 34, 997–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Maeda, T.; Grant, S.; Grant, G.; Sporns, P. Confirmation of fructans biosynthesized in vitro from 1-C-13 glucose in asparagus tissues using MALDI-TOF MS and ESI-MS. J. Plant Physiol. 2013, 170, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Maeda, T.; Nomura, S.; Suzuki, M.; Grant, G.; Sporns, P. Rapid analysis of fructans and comparison of fructan profiles in several different types of asparagus storage roots using MALDI-TOF MS. J. Horticult. Sci. Biotechnol. 2011, 86, 210–216. [Google Scholar] [CrossRef]
- Hisano, H.; Kanazawa, A.; Yoshida, M.; Humphreys, M.O.; Iizuka, M.; Kitamura, K.; Yamada, T. Coordinated expression of functionally diverse fructosyltransferase genes is associated with fructan accumulation in response to low temperature in perennial ryegrass. New Phytol. 2008, 178, 766–780. [Google Scholar] [CrossRef] [PubMed]
- Eigner, W.D.; Abuja, P.; Beck, R.H.F.; Praznik, W. Physicochemical characterization of inulin and sinistrin. Carbohydr. Res. 1988, 180, 87–95. [Google Scholar] [CrossRef]
- Ponce, J.; Macías, E.; Soltero, J.; Fernández, V.; Zúñiga, V.; Escalona, H. Physical-Chemical and non-linear rheological properties of aqueous solutions of agave fructans. e-Gnosis 2008, 6, 1–23. [Google Scholar]
- Wolff, D.; Czapla, S.; Heyer, A.G.; Radosta, S.; Mischnick, P.; Springer, J. Globular shape of high molar mass inulin revealed by static light scattering and viscometry. Polymer 2000, 41, 8009–8016. [Google Scholar] [CrossRef]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P.N. High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev. Cell 2016, 39, 508–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shulse, C.N.; Cole, B.J.; Ciobanu, D.; Lin, J.; Yoshinaga, Y.; Gouran, M.; Turco, G.M.; Zhu, Y.; O’Malley, R.C.; Brady, S.M.; et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019, 27, 2241–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.Q.; Xu, Z.G.; Shang, G.D.; Wang, J.W. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol. Plant 2019, 12, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Jean-Baptiste, K.; McFaline-Figueroa, J.L.; Alexandre, C.M.; Dorrity, M.W.; Saunders, L.; Bubb, K.L.; Trapnell, C.; Fields, S.; Queitsch, C.; Cuperus, J.T. Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell 2019, 31, 993–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, K.H.; Huang, L.; Kang, H.M.; Schiefelbein, J. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019, 179, 1444–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.L.; Li, H.; Bhatti, S.; Zhou, S.P.; Yang, Y.; Fish, T.; Thannhauser, T.W. The Al-induced proteomes of epidermal and outer cortical cells in root apex of cherry tomato ‘LA 2710’. J. Proteom. 2020, 211, 11. [Google Scholar] [CrossRef]
- Dembinsky, D.; Woll, K.; Saleem, M.; Liu, Y.; Fu, Y.; Borsuk, L.A.; Lamkemeyer, T.; Fladerer, C.; Madlung, J.; Barbazuk, B.; et al. Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 2007, 145, 575–588. [Google Scholar] [CrossRef] [Green Version]
- Lemoine, R. Sucrose transporters in plants: Update on function and structure. Biochim. Biophys. Acta-Biomembr. 2000, 1465, 246–262. [Google Scholar] [CrossRef] [Green Version]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source-sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef] [Green Version]
- Münch, E. Die Stoffbewegungen in der Pflanze; Verlag von Gustav Fischer: Jena, Germany, 1930. [Google Scholar]
- de Dios, E.A.; Vargas, A.G.; Santos, M.L.D.; Simpson, J. New insights into plant glycoside hydrolase family 32 in Agave species. Front. Plant Sci. 2015, 6, 14. [Google Scholar]







| Peak # | RT [min] | Compound | DP | Molecular Family/Fructan-Type | [M + K]+ |
|---|---|---|---|---|---|
| 1 | 3.24 | Glucose | 1 | Monosaccharide | |
| 2 | 3.56 | Fructose | 1 | Monosaccharide | |
| 3 | 4.08 | Melibiose | 2 | Disaccharide | 381 |
| 4 | 5.49 | Sucrose | 2 | Disaccharide | 381 |
| 5 | 9.16 | Raffinose | 3 | Raffinose-type oligosaccharides | 543 |
| 6 | 9.84 | 1-Kestose | 3 | Inulin-type fructan | 543 |
| 7 | 10.35 | Unknown | |||
| 8 | 10.58 | Maltose | 2 | Maltose-type oligosaccharides | |
| 9 | 10.93 | Unknown | |||
| 10 | 11.59 | Unknown | |||
| 11 | 11.94 | Unknown | |||
| 12 | 12.51 | 6G-Kestose (NS1-DP3) | 3 | Neoseries (NS)1-type fructan | 543 |
| 13 | 13.35 | Nystose (1,1-Kestotetraose, KT) (Inulin-DP4) | 4 | Inulin-type fructan | 705 |
| 14 | 14.20 | 1&6G-KT (NS2-DP4) | 4 | NS2-type fructan | 705 |
| 15 | 15.40 | Unknown | |||
| 16 | 15.83 | 1, 6G-KT (NS1-DP4) | 4 | NS1-type fructan | 705 |
| 17 | 16.45 | 1,1,1-Kestopentaose (KP) (Inulin-DP5) | 5 | Inulin-type fructan | 867 |
| 18 | 17.13 | 1,1&6G-KP (NS2-DP5), 1&1, 6G-KP (NS3-DP5) | 5 | NS2&NS3-type fructan | 867 |
| 19 | 17.69 | Unknown | |||
| 20 | 18.04 | Unknown | |||
| 21 | 18.33 | Unknown | |||
| 22 | 18.66 | Unknown | |||
| 23 | 19.03 | 1,1, 6G-KP (NS1-DP5) | 5 | NS1-type fructan | 867 |
| 24 | 19.43 | 1,1,1,1-Kestohexaose (KH) (Inulin-DP6) | 6 | Inulin-type fructan | 1029 |
| 25 | 20.04 | 1,1,1&6G-KH (NS2-DP6), 1&1,1, 6G-KH (NS3-DP6), 1,1&1, 6G-KH (NS4-DP6) | 6 | NS2-4-type fructan | 1029 |
| 26 | 20.89 | Unknown | |||
| 27 | 21.17 | Unknown | |||
| 28 | 21.71 | Unknown | |||
| 29 | 22.11 | 1,1,1, 6G-KP (NS1-DP6) and Inulin-DP7 | 6&7 | Inulin & NS1-type fructan | 1029&1191 |
| 30 | 22.92 | 1,1,1,1&6G-KH (NS2-DP7), 1&1,1,1, 6G-KH (NS3-DP7), 1,1,1&1, 6G-KH (NS4-DP7) | 7 | NS2-4-type fructan | 1191 |
| 31 | 23.42 | Unknown | |||
| 32 | 23.88 | Unknown | |||
| 33 | 24.23 | Unknown | |||
| 34 | 24.95 | NS1-DP7 and Inulin-DP8 | 7&8 | Inulin & NS1-type fructan | 1191&1353 |
| 35 | 25.64 | NS2-DP8, NS3-DP8, NS4-DP8 | 8 | NS2-4-type fructan | 1353 |
| 36 | 26.50 | Unknown | |||
| 37 | 26.84 | Unknown | |||
| 38 | 27.65 | NS1-DP8 and Inulin-DP9 | 8&9 | Inulin & NS1-type fructan | 1353&1515 |
| 39 | 28.04 | NS2-DP9, NS3-DP9, NS4-DP9 | 9 | NS2-4-type fructan | 1515 |
| 40 | 28.93 | Unknown | |||
| 41 | 29.32 | Unknown | |||
| 42 | 30.22 | NS1-DP9, Inulin-DP10, NS2-DP10, NS3-DP10, NS4-DP10 | 9&10 | Inulin & NS1-NS4-type fructan | 1515&1677 |
| 43 | 31.13 | Unknown | |||
| 44 | 32.33 | NS1-DP10, Inulin-DP11, NS2-DP11, NS3-DP11, NS4-DP11 | 10&11 | Inulin & NS1-NS4-type fructan | 1677&1839 |
| 45 | 33.13 | Unknown | |||
| 46 | 34.27 | NS1-DP11, Inulin-DP12, NS2-DP12, NS3-DP12, NS4-DP12 | 11&12 | Inulin & NS1-NS4-type fructan | |
| 47 | 35.02 | Unknown | |||
| 48 | 36.08 | NS1-DP12, Inulin-DP13, NS2-DP13, NS3-DP13, NS4-DP13 | 12&13 | Inulin & NS1-NS4-type fructan | |
| 49 | 36.78 | Unknown | |||
| 50 | 37.77 | NS1-DP13, Inulin-DP14, NS2-DP14, NS3-DP14, NS4-DP14 | 13&14 | Inulin & NS1-NS4-type fructan | |
| 51 | 38.42 | Unknown | |||
| 52 | 39.33 | NS1-DP14, Inulin-DP15, NS2-DP15, NS3-DP15, NS4-DP15 | 14&15 | Inulin & NS1-NS4-type fructan | |
| 53 | 39.93 | Unknown | |||
| 54 | 40.81 | NS1-DP15, Inulin-DP16, NS2-DP16, NS3-DP16, NS4-DP16 | 15&16 | Inulin & NS1-NS4-type fructan | |
| 55 | 41.42 | Unknown | |||
| 56 | 42.20 | NS1-DP16, Inulin-DP17, NS2-DP17, NS3-DP17, NS4-DP17 | 16&17 | Inulin & NS1-NS4-type fructan | |
| 57 | 42.81 | Unknown |
| Peak | 13 | 14 | 16 | 17 | 18 | 23 | 24 | 25 | 29 | 30 | 34 | 35 | 38 | 39 | 42 | 44 | 46 | 48 | 50 | 52 | 54 | 56 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Outer tissues | 1-Kestose | 1.4 | 1.2 | 1.1 | 1.7 | 0.8 | 1.8 | 4.7 | 1.0 | 1.6 | 0.9 | 2.7 | 1.0 | 11.0 | 1.7 | 1.3 | 1.9 | 3.4 | 6.4 | 13.0 | 28.8 | 61.7 | 132.0 |
| 6G-Kestose | 1.1 | 1.0 | 0.9 | 1.4 | 0.7 | 1.5 | 3.8 | 0.8 | 1.3 | 0.7 | 2.2 | 0.8 | 8.8 | 1.3 | 1.1 | 1.6 | 2.7 | 5.2 | 10.5 | 23.2 | 49.6 | 106.2 | |
| Middle tissues | 1-Kestose | 1.4 | 1.1 | 1.0 | 1.5 | 0.8 | 1.7 | 3.4 | 0.8 | 1.3 | 0.8 | 1.9 | 0.9 | 7.6 | 1.5 | 0.9 | 1.2 | 1.7 | 2.6 | 4.4 | 8.0 | 15.2 | 29.3 |
| 6G-Kestose | 1.3 | 1.0 | 0.9 | 1.3 | 0.7 | 1.5 | 3.0 | 0.7 | 1.2 | 0.7 | 1.7 | 0.8 | 6.7 | 1.3 | 0.8 | 1.0 | 1.5 | 2.3 | 3.9 | 7.1 | 13.4 | 25.9 | |
| Inner Tissues | 1-Kestose | 1.5 | 1.0 | 0.9 | 1.8 | 0.6 | 1.2 | 4.5 | 0.6 | 1.1 | 0.5 | 1.9 | 0.6 | 6.3 | 0.9 | 0.8 | 1.3 | 2.1 | 3.7 | 6.7 | 13.0 | 25.2 | 48.7 |
| 6G-Kestose | 1.5 | 1.0 | 0.9 | 1.8 | 0.6 | 1.2 | 4.5 | 0.6 | 1.2 | 0.5 | 1.9 | 0.6 | 6.3 | 0.9 | 0.8 | 1.3 | 2.1 | 2.8 | 7.0 | 13.3 | 25.6 | 49.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witzel, K.; Matros, A. Fructans Are Differentially Distributed in Root Tissues of Asparagus. Cells 2020, 9, 1943. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9091943
Witzel K, Matros A. Fructans Are Differentially Distributed in Root Tissues of Asparagus. Cells. 2020; 9(9):1943. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9091943
Chicago/Turabian StyleWitzel, Katja, and Andrea Matros. 2020. "Fructans Are Differentially Distributed in Root Tissues of Asparagus" Cells 9, no. 9: 1943. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9091943