Principal Postulates of Centrosomal Biology. Version 2020
Abstract
:1. Introduction
2. Postulates of Centrosomal Biology
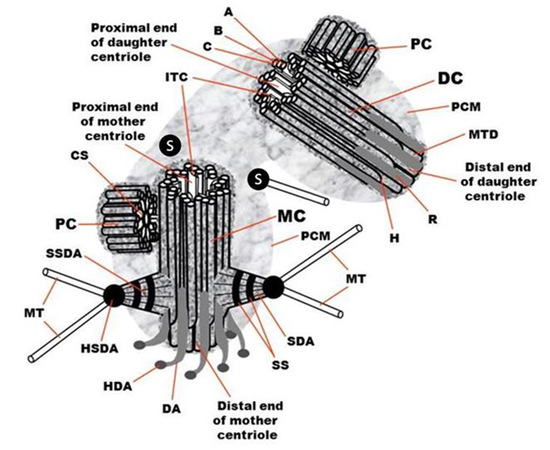
2.1. The Centriole and the Basal Body Are Two Forms of the Same Organelle
2.2. The Centriole Is a Polar Structure with Two Morphologically and Functionally Different Ends
2.3. The Outer Diameter of the Distal Part of the Centriolar Cylinder Is Smaller Than That of the Proximal End
2.4. The Centriolar Cylinder Length Is Highly Regulated
2.5. In the Centrosome of Proliferating Cells, Two Centrioles Differ Structurally and Functionally
2.6. The Proximal Ends of the Mother and Daughter Centrioles Are Connected via a Bundle of Thin Fibers
2.7. The Centriole Lumen Helps in Stabilizing the Centriole
2.8. Maintaining a Cylindrical Shape of Centrioles Can Be Independent of MT Triplets
2.9. The Structure and Activity of Centrosomes Differ in Interphase and Mitotic Cells
2.10. New Centrioles Are Usually Formed in Association with Mother Centrioles but Can Be Formed without Preexisting Centriole De Novo
2.11. The Centrosomes Have Four Types of MT Nucleating Activity
- (1)
- Formation of two types of interphase MTs: (i) a radial MTs system around the centriole; and (ii) non-centrosomal (free) cytoplasmic MTs that were polymerized on the centrosome and later released to the cytoplasm.
- (2)
- Formation of three types of mitotic spindle microtubules: (i) astral MTs; (ii) kinetochore MTs; and (iii) interzonal MTs.
- (3)
- Formation of MT of procentrioles.
- (4)
2.12. The Complete Process of Centrioles Maturation from Procentriole to Mother Centriole Takes More Than One and a Half Cell Cycles in Duration
2.13. The Centrosome Is the Center of the Organization of Actin Microfilaments in the Cell
2.14. Centrioles Can Gain an Atypical Structure and Composition and Become Undetected Using Standard Expectations and Techniques
2.15. The Centrosome Is a Polyfunctional, Multi-Protein, and Cell Regulation Complex; Some of Its Proteins Simultaneously Regulate Several Intracellular Processes
3. Questions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flemming, W. Studien in der entwicklungsgeschichte der najaden. Sitz. Akad. Wissensch Wien. 1875, 71, 81–147. [Google Scholar]
- Hertwig, O. Beitrage zur kenntniss der bildung, befruchtung und theilung des thierischen eies. Morphol. Jb. 1875, 1, 347–434. [Google Scholar] [CrossRef]
- Van Beneden, E. Recherches sur les dicyémides, survivants actuels d’un embranchement des mésozoaires. Bull. Acad. Roy. Méd. Belg. 1876, 41, 1160–1205. [Google Scholar]
- Van Beneden, E.; Neyt, A. Nouvelles recherches sur la fecondation et la division mitoshes l’ascaride megalocephale. Bull. Acad. R. Méd. Belg. 1887, 14, 1–81. Available online: https://orbi.uliege.be/handle/2268/160175 (accessed on 23 September 2020).
- Boveri, T. Die Bildung der Richtungskörper bei Ascaris megalocephala und Ascaris lumbricoides. Zellen-Studien Heft 1; Verlag von Gustav Fischer: Jena, Germany, 1887; Available online: https://www.biozentrum.uni-wuerzburg.de/zeb/research/topics/theodor-boveri/cell-studies/ (accessed on 23 September 2020).
- Boveri, T. Ueber das Verhalten der Centrosomen bei der Befruchtung des Seeigel-Eies, nebst allgemeinen Bemerkungen über Centrosomen und Verwandtes. Verhandl. Phys.-Med. Ges. Würzburg. 1895, 29, 1–75. Available online: https://www.biozentrum.uni-wuerzburg.de/fileadmin/07020100/2018/Downloads_PDF/Boveri_1895a.pdf (accessed on 23 September 2020).
- Henneguy, L.F. Sur les rapports des cils vibratiles avec les centrosomes. Arch. Microsc. Morph. Exp. 1898, 1, 481–496. [Google Scholar]
- Von Lenhossék, M. Uber flimmerzellen. Verh. Anat. Ges. Kiel. 1898, 12, 106–128. [Google Scholar]
- Afzelius, B. Electron microscopy of the sperm tail; results obtained with a new fixative. J. Biophys. Biochem. Cytol. 1959, 5, 269–278. [Google Scholar] [CrossRef]
- Gibbons, I.R.; Grimstone, A.V. On flagellar structure in certain flagellates. J. Biophys. Biochem. Cytol. 1960, 7, 697–716. [Google Scholar] [CrossRef] [Green Version]
- Brinkley, B.R.; Stubblefield, E. Ultrastructure and interaction of the kinetochore and centriole in mitosis and meiosis. In Advances in Cell Biology; Prescott, D.M., Mc Conkey, E., Eds.; Appleton-Century Crofts: New York, NY, USA, 1970; Volume 1, pp. 119–184. [Google Scholar]
- Uzbekov, R.; Prigent, C. Clockwise or anticlockwise? Turning the centriole triplets in the right direction! FEBS Lett. 2007, 581, 1251–1254. [Google Scholar] [CrossRef] [Green Version]
- Uzbekov, R.E.; Alieva, I.B. Centrosome: History of Study and New Discoveries. From Cytoplasmic Granule to the Center of Intracellular Regulation; Moscow University Press: Moscow, Russia, 2013. [Google Scholar]
- Sulston, J.E.; Albertson, D.G.; Thomson, J.N. The caenorhabditis elegans male: Postembryonic development of nongonadal structures. Dev. Biol. 1980, 78, 542–576. [Google Scholar] [CrossRef]
- Riparbelli, M.G.; Dallai, R.; Callaini, G. The insect centriole: A land of discovery. Tissue Cell 2010, 42, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, M.; Callaini, G.; Riparbelli, M.G. The drosophila centriole–conversion of doublets into triplets within the stem cell niche. J. Cell Sci. 2015, 128, 2437–2442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favard, P. Evolution des ultrastructures cellulaires au cours de la spermatogenese de l’ascaris (ascaris megalocephala, schrank = parascaris equorum, goerze). Ann. Des Sci. Nat. Zool. Et Biol. Anim. 1961, 12, 52–152. [Google Scholar]
- Wolf, N.; Hirsh, D.; McIntosh, J.R. Spermatogenesis in males of the free-living nematode, caenorhabditis elegans. J. Ultrastruct. Res. 1978, 63, 155–169. [Google Scholar] [CrossRef]
- Pelletier, L.; O’Toole, E.; Schwager, A.; Hyman, A.A.; Muller-Reichert, T. Centriole assembly in caenorhabditis elegans. Nature 2006, 444, 619–623. [Google Scholar] [CrossRef]
- Uzbekov, R.; Garanina, A.; Bressac, C. Centrioles without microtubules: A new morphological type of centriole. Biol. Open 2018, 7, bio036012. [Google Scholar] [CrossRef] [Green Version]
- Uzbekov, R.; Garanina, A.S.; Burlaud-Gaillard, J.; Bressac, C. The flagellum of the shortest spermatozoon in the animal kingdom. Elongation and shortening of the axoneme in the process of spermiogenesis of the parasitic wasp cotesia congregata. In Flagella and Cilia: Types, Structure and Functions; Uzbekov, R., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2018; pp. 83–108. [Google Scholar]
- McNitt, R. Centriole ultrastructure and its possible role in microtubule formation in an aquatic fungus. Protoplasma 1974, 80, 91–108. [Google Scholar] [CrossRef]
- Greenan, G.A.; Keszthelyi, B.; Vale, R.D.; Agard, D.A. Insights into centriole geometry revealed by cryotomography of doublet and triplet centrioles. ELife 2018, 7, e36851. [Google Scholar] [CrossRef]
- Jana, S.C.; Girotra, M.; Ray, K. Heterotrimeric kinesin-ii is necessary and sufficient to promote different stepwise assembly of morphologically distinct bipartite cilia in drosophila antenna. Mol. Biol. Cell 2011, 22, 769–781. [Google Scholar] [CrossRef]
- Gottardo, M.; Pollarolo, G.; Llamazares, S.; Reina, J.; Riparbelli, M.G.; Callaini, G.; Gonzalez, C. Loss of centrobin enables daughter centrioles to form sensory cilia in drosophila. Curr. Biol. 2015, 25, 2319–2324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nechipurenko, I.V.; Berciu, C.; Sengupta, P.; Nicastro, D. Centriolar remodeling underlies basal body maturation during ciliogenesis in caenorhabditis elegans. ELife 2017, 6, e25686. [Google Scholar] [CrossRef] [PubMed]
- Ringo, D.L. The arrangement of subunits in flagellar fibers. J. Ultrastruct. Res. 1967, 17, 266–277. [Google Scholar] [CrossRef]
- Tilney, L.G.; Bryan, J.; Bush, D.J.; Fujiwara, K.; Mooseker, M.S.; Murphy, D.B.; Snyder, D.H. Microtubules: Evidence for 13 protofilaments. J. Cell Biol. 1973, 59, 267–275. [Google Scholar] [CrossRef]
- André, J. Le centriole et la région centrosomienne. J. Microsc. 1964, 3, 1–23. [Google Scholar]
- Wheatley, D.N. The Centriole: A Central Enigma of Cell Biology; Elsevier Biomedical Press: Amsterdam, The Netherlands; New York, NY, USA, 1982. [Google Scholar]
- Ou, Y.; Rattner, J.B. The centrosome in higher organisms: Structure, composition, and duplication. Int. Rev. Cytol. 2004, 238, 119–182. [Google Scholar]
- Guichard, P.; Hachet, V.; Majubu, N.; Neves, A.; Demurtas, D.; Olieric, N.; Fluckiger, I.; Yamada, A.; Kihara, K.; Nishida, Y.; et al. Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr. Biol. 2013, 23, 1620–1628. [Google Scholar] [CrossRef] [Green Version]
- Gall, J.G. Centriole replication. A study of spermatogenesis in the snail viviparus. J. Biophys. Biochem. Cytol. 1961, 10, 163–193. [Google Scholar] [CrossRef] [Green Version]
- Kitagawa, D.; Vakonakis, I.; Olieric, N.; Hilbert, M.; Keller, D.; Olieric, V.; Bortfeld, M.; Erat, M.C.; Fluckiger, I.; Gonczy, P.; et al. Structural basis of the 9-fold symmetry of centrioles. Cell 2011, 144, 364–375. [Google Scholar] [CrossRef] [Green Version]
- Van Breugel, M.; Hirono, M.; Andreeva, A.; Yanagisawa, H.A.; Yamaguchi, S.; Nakazawa, Y.; Morgner, N.; Petrovich, M.; Ebong, I.O.; Robinson, C.V.; et al. Structures of sas-6 suggest its organization in centrioles. Science 2011, 331, 1196–1199. [Google Scholar] [CrossRef] [Green Version]
- Vorobjev, I.A.; Chentsov, Y.S. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982, 93, 938–949. [Google Scholar] [CrossRef]
- Friedlander, M.; Wahrman, J. Giant centrioles in neuropteran meiosis. J. Cell Sci. 1966, 1, 129–144. [Google Scholar] [PubMed]
- Spektor, A.; Tsang, W.Y.; Khoo, D.; Dynlacht, B.D. Cep97 and cp110 suppress a cilia assembly program. Cell 2007, 130, 678–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szollosi, D. The structure and function of centrioles and their satellites in the jellyfish phialidium gregarium. J. Cell Biol. 1964, 21, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.G. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J. Cell Biol. 1972, 54, 246–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhard, W.; de Harven, E. L’ultrastructure du centriole et d’autres éléments de l’appareil achromatique. In Vierter Internationaler Kongress fur Elektronenmikroskopie, 1958; Bargmann, W., Möllenstedt, G., Niehrs, H., Peters, D., Ruska, E., Wolpers, C., Eds.; Springer: Berlin, Germany, 1960; Volume 2, pp. 217–227. [Google Scholar]
- Stubblefield, E.B.; Brinkley, R. Architecture and function of the mammalian centriole. In Formation and Fate of Cell Organelles; Warren, K.B., Ed.; Academic Press Inc.: New York, NY, USA, 1967; pp. 175–218. [Google Scholar]
- Vorobjev, I.A.; Chentsov, Y.S. The ultrastructure of the centrosome in the cells of hematopoietic tissues of axolotl. Tsitologiia 1977, 19, 598–603. [Google Scholar]
- Uzbekov, R.; Alieva, I. Who are you, subdistal appendages of centriole? Open Biol. 2018, 8, 180062. [Google Scholar] [CrossRef] [Green Version]
- Doolin, P.F.; Birge, W.J. Ultrastructural organization of cilia and basal bodies of the epithelium of the choroid plexus in the chick embryo. J. Cell Biol. 1966, 29, 333–345. [Google Scholar] [CrossRef]
- Sorokin, S.P. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 1968, 3, 207–230. [Google Scholar]
- Wolfe, J. Basal body fine structure and chemistry. Adv. Cell Mol. Biol. 1972, 2, 151–192. [Google Scholar]
- Gibbons, I.R. The relationship between the fine structure and direction of beat in gill cilia of a lamellibranch mollusc. J. Biophys. Biochem. Cytol. 1961, 11, 179–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, Y.; Rattner, J.B. A subset of centrosomal proteins are arranged in a tubular conformation that is reproduced during centrosome duplication. Cell Motil. Cytoskelet. 2000, 47, 13–24. [Google Scholar] [CrossRef]
- Guichard, P.; Chretien, D.; Marco, S.; Tassin, A.M. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010, 29, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Hara, P.T. Spiral tilt of triplet fibers in human leukocyte centrioles. J. Ultrastruct. Res. 1970, 31, 195–198. [Google Scholar] [CrossRef]
- Le Guennec, M.; Klena, N.; Gambarotto, D.; Laporte, M.H.; Tassin, A.M.; van den Hoek, H.; Erdmann, P.S.; Schaffer, M.; Kovacik, L.; Borgers, S.; et al. A helical inner scaffold provides a structural basis for centriole cohesion. Sci. Adv. 2020, 6, eaaz4137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avidor-Reiss, T.; Gopalakrishnan, J. Cell cycle regulation of the centrosome and cilium. Drug Discov. Today. Dis. Mech. 2013, 10, e119–e124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basiri, M.L.; Blachon, S.; Chim, Y.C.; Avidor-Reiss, T. Imaging centrosomes in fly testes. J. Vis. Exp. 2013, e50938. [Google Scholar] [CrossRef]
- Schreiner, A.; Schreiner, K.E. Űber die entwicklung der männlichen geschlechtszellen von myxine glutinosa (l.). Arch. Biol. 1905, 21, 183–357. [Google Scholar]
- Guichard, P.; Desfosses, A.; Maheshwari, A.; Hachet, V.; Dietrich, C.; Brune, A.; Ishikawa, T.; Sachse, C.; Gonczy, P. Cartwheel architecture of trichonympha basal body. Science 2012, 337, 553. [Google Scholar] [CrossRef]
- Azimzadeh, J.; Hergert, P.; Delouvee, A.; Euteneuer, U.; Formstecher, E.; Khodjakov, A.; Bornens, M. Hpoc5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 2009, 185, 101–114. [Google Scholar] [CrossRef] [Green Version]
- Blachon, S.; Cai, X.; Roberts, K.A.; Yang, K.; Polyanovsky, A.; Church, A.; Avidor-Reiss, T. A proximal centriole-like structure is present in drosophila spermatids and can serve as a model to study centriole duplication. Genetics 2009, 182, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohlmaier, G.; Loncarek, J.; Meng, X.; McEwen, B.F.; Mogensen, M.M.; Spektor, A.; Dynlacht, B.D.; Khodjakov, A.; Gonczy, P. Overly long centrioles and defective cell division upon excess of the sas-4-related protein cpap. Curr. Biol. 2009, 19, 1012–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.J.; Fu, R.H.; Wu, K.S.; Hsu, W.B.; Tang, T.K. Cpap is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 2009, 11, 825–831. [Google Scholar] [CrossRef]
- Schmidt, T.I.; Kleylein-Sohn, J.; Westendorf, J.; Le Clech, M.; Lavoie, S.B.; Stierhof, Y.D.; Nigg, E.A. Control of centriole length by cpap and cp110. Curr. Biol. 2009, 19, 1005–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comartin, D.; Gupta, G.D.; Fussner, E.; Coyaud, E.; Hasegan, M.; Archinti, M.; Cheung, S.W.; Pinchev, D.; Lawo, S.; Raught, B.; et al. Cep120 and spice1 cooperate with cpap in centriole elongation. Curr. Biol. 2013, 23, 1360–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.N.; Wu, C.T.; Lin, Y.C.; Hsu, W.B.; Tang, C.J.; Chang, C.W.; Tang, T.K. Cep120 interacts with cpap and positively regulates centriole elongation. J. Cell Biol. 2013, 202, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Kong, D.; Sahabandu, N.; Sullenberger, C.; Vasquez-Limeta, A.; Luvsanjav, D.; Lukasik, K.; Loncarek, J. Prolonged mitosis results in structurally aberrant and over-elongated centrioles. J. Cell Biol. 2020, 219, e201910019. [Google Scholar] [CrossRef] [Green Version]
- Oakley, C.E.; Oakley, B.R. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipa gene of aspergillus nidulans. Nature 1989, 338, 662–664. [Google Scholar] [CrossRef]
- Joshi, H.C.; Palacios, M.J.; McNamara, L.; Cleveland, D.W. Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature 1992, 356, 80–83. [Google Scholar] [CrossRef]
- Zheng, Y.; Wong, M.L.; Alberts, B.; Mitchison, T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 1995, 378, 578–583. [Google Scholar] [CrossRef]
- Moritz, M.; Zheng, Y.; Alberts, B.M.; Oegema, K. Recruitment of the gamma-tubulin ring complex to drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998, 142, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Oegema, K.; Wiese, C.; Martin, O.C.; Milligan, R.A.; Iwamatsu, A.; Mitchison, T.J.; Zheng, Y. Characterization of two related drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 1999, 144, 721–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, X.; Habedanck, R.; Nigg, E.A. A complex of two centrosomal proteins, cap350 and fop, cooperates with eb1 in microtubule anchoring. Mol. Biol. Cell 2006, 17, 634–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.C.; Badano, J.L.; Sibold, S.; Esmail, M.A.; Hill, J.; Hoskins, B.E.; Leitch, C.C.; Venner, K.; Ansley, S.J.; Ross, A.J.; et al. The bardet-biedl protein bbs4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004, 36, 462–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimzadeh, J.; Bornens, M. Structure and duplication of the centrosome. J. Cell Sci. 2007, 120, 2139–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Harven, E. The centriole and the mitotic spindle. In The Nucleus; Dalton, A.J., Haguenau, F., Eds.; Academic Press: New York, NY, USA, 1968; pp. 197–227. [Google Scholar]
- Tilney, L.G.; Goddard, J. Nucleated sites for the assembly of cytoplasmic microtubules in the ectodermal cells of blastulae of arbacia punctulata. J. Cell Biol. 1970, 46, 564–575. [Google Scholar] [CrossRef] [Green Version]
- Dammermann, A.; Merdes, A. Assembly of centrosomal proteins and microtubule organization depends on pcm-1. J. Cell Biol. 2002, 159, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Kubo, A.; Tsukita, S. Non-membranous granular organelle consisting of pcm-1: Subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 2003, 116, 919–928. [Google Scholar] [CrossRef] [Green Version]
- Conkar, D.; Bayraktar, H.; Firat-Karalar, E.N. Centrosomal and ciliary targeting of ccdc66 requires cooperative action of centriolar satellites, microtubules and molecular motors. Sci. Rep. 2019, 9, 14250. [Google Scholar] [CrossRef] [Green Version]
- Prosser, S.L.; Pelletier, L. Centriolar satellite biogenesis and function in vertebrate cells. J. Cell Sci. 2020, 133, jcs239566. [Google Scholar] [CrossRef]
- Bornens, M.; Paintrand, M.; Berges, J.; Marty, M.C.; Karsenti, E. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskelet. 1987, 8, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Tournier, F.; Komesli, S.; Paintrand, M.; Job, D.; Bornens, M. The intercentriolar linkage is critical for the ability of heterologous centrosomes to induce parthenogenesis in xenopus. J. Cell Biol. 1991, 113, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Paintrand, M.; Moudjou, M.; Delacroix, H.; Bornens, M. Centrosome organization and centriole architecture: Their sensitivity to divalent cations. J. Struct. Biol. 1992, 108, 107–128. [Google Scholar] [CrossRef]
- Yang, J.; Liu, X.; Yue, G.; Adamian, M.; Bulgakov, O.; Li, T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 2002, 159, 431–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahe, S.; Stierhof, Y.D.; Wilkinson, C.J.; Leiss, F.; Nigg, E.A. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005, 171, 27–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahmanyar, S.; Kaplan, D.D.; Deluca, J.G.; Giddings, T.H., Jr.; O’Toole, E.T.; Winey, M.; Salmon, E.D.; Casey, P.J.; Nelson, W.J.; Barth, A.I. Beta-catenin is a nek2 substrate involved in centrosome separation. Genes Dev. 2008, 22, 91–105. [Google Scholar] [CrossRef] [Green Version]
- Tsou, M.F.; Stearns, T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 2006, 442, 947–951. [Google Scholar] [CrossRef]
- Uzbekov, R.; Kireyev, I.; Prigent, C. Centrosome separation: Respective role of microtubules and actin filaments. Biol. Cell 2002, 94, 275–288. [Google Scholar] [CrossRef]
- Glover, D.M.; Leibowitz, M.H.; McLean, D.A.; Parry, H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 1995, 81, 95–105. [Google Scholar] [CrossRef] [Green Version]
- Roghi, C.; Giet, R.; Uzbekov, R.; Morin, N.; Chartrain, I.; Le Guellec, R.; Couturier, A.; Doree, M.; Philippe, M.; Prigent, C. The xenopus protein kinase peg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell Sci. 1998, 111, 557–572. [Google Scholar] [PubMed]
- Giet, R.; Uzbekov, R.; Cubizolles, F.; Le Guellec, K.; Prigent, C. The xenopus laevis aurora-related protein kinase peg2 associates with and phosphorylates the kinesin-related protein xleg5. J. Biol. Chem. 1999, 274, 15005–15013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowley, D.O.; Rivera-Perez, J.A.; Schliekelman, M.; He, Y.J.; Oliver, T.G.; Lu, L.; O’Quinn, R.; Salmon, E.D.; Magnuson, T.; Van Dyke, T. Aurora-a kinase is essential for bipolar spindle formation and early development. Mol. Cell. Biol. 2009, 29, 1059–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blangy, A.; Arnaud, L.; Nigg, E.A. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor hseg5 to the dynactin subunit p150. J. Biol. Chem. 1997, 272, 19418–19424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, H.A.; Nigg, E.A. Cell-cycle control: Polo-like kinases join the outer circle. Trends Cell Biol. 1997, 7, 63–68. [Google Scholar] [CrossRef]
- Glover, D.M.; Hagan, I.M.; Tavares, A.A. Polo-like kinases: A team that plays throughout mitosis. Genes Dev. 1998, 12, 3777–3787. [Google Scholar] [CrossRef] [Green Version]
- Golsteyn, R.M.; Lane, H.A.; Mundt, K.E.; Arnaud, L.; Nigg, E.A. The family of polo-like kinases. Prog. Cell Cycle Res. 1996, 2, 107–114. [Google Scholar] [PubMed]
- Gaglio, T.; Saredi, A.; Bingham, J.B.; Hasbani, M.J.; Gill, S.R.; Schroer, T.A.; Compton, D.A. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 1996, 135, 399–414. [Google Scholar] [CrossRef]
- Gaglio, T.D.; Dionne, M.A.; Compton, D.A. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 1997, 138, 1055–1066. [Google Scholar] [CrossRef] [Green Version]
- Echeverri, C.J.P.; Paschal, B.M.; Vaughan, K.T.; Vallee, R.B. Molecular characterization of the 50-kd subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 1996, 132, 617–633. [Google Scholar] [CrossRef] [Green Version]
- Boleti, H.K.; Karsenti, E.; Vernos, I. Xklp2, a novel xenopus centrosomal kinesin-like protein required for centrosome sepqration during mitosis. Cell 1996, 84, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Sawin, K.E.; LeGuellec, K.; Philippe, M.; Mitchison, T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 1992, 359, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Verma, S.; Mitchison, T.J. Xctk2: A kinesin-related protein that promotes mitotic spindle assembly in xenopus laevis egg extracts. J. Cell Biol. 1997, 136, 859–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Harven, E.; Bernhard, W. Etude au microscope électronique de l’ultrastructure du centriole chez les vertébrés. Z. Zellforsch. U. Mokr. Anat. 1956, 45, 1–378. [Google Scholar]
- Osborn, M.; Weber, K. Cytoplasmic microtubules in tissue culture cells appear to grow from an organizing structure towards the plasma membrane. Proc. Natl. Acad. Sci. USA 1976, 73, 867–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, C.G.; Jensen, L.C.; Rieder, C.L. The occurrence and structure of primary cilia in a subline of potorous tridactylus. Exp. Cell Res. 1979, 123, 444–449. [Google Scholar] [CrossRef]
- Fais, D.A.; Nadezhdina, E.S.; Chentsov, Y.S. The centriolar rim. The structure that maintains the configuration of centrioles and basal bodies in the absence of their microtubules. Exp. Cell Res. 1986, 164, 27–34. [Google Scholar] [CrossRef]
- Steffen, W.; Linck, R.W. Evidence for tektins in centrioles and axonemal microtubules. Proc. Natl. Acad. Sci. USA 1988, 85, 2643–2647. [Google Scholar] [CrossRef] [Green Version]
- Stephens, R.E.; Lemieux, N.A. Tektins as structural determinants in basal bodies. Cell Motil. Cytoskelet. 1998, 40, 379–392. [Google Scholar] [CrossRef]
- Plessmann, U.; Weber, K. Mammalian sperm tubulin: An exceptionally large number of variants based on several posttranslational modifications. J. Protein Chem. 1997, 16, 385–390. [Google Scholar] [CrossRef]
- Bobinnec, Y.; Khodjakov, A.; Mir, L.M.; Rieder, C.L.; Edde, B.; Bornens, M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 1998, 143, 1575–1589. [Google Scholar] [CrossRef] [Green Version]
- Steib, E.; Gambarotto, D.; Laporte, M.H.; Olieric, N.; Zheng, C.; Borgers, S.; Olieric, V.; Le Guennec, M.; Koll, F.; Tassin, A.-M.; et al. The inner scaffold protects from centriole fracture. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Pearson, C.G.; Osborn, D.P.; Giddings, T.H., Jr.; Beales, P.L.; Winey, M. Basal body stability and ciliogenesis requires the conserved component poc1. J. Cell Biol. 2009, 187, 905–920. [Google Scholar] [CrossRef] [Green Version]
- Lattao, R.; Kovacs, L.; Glover, D.M. The centrioles, centrosomes, basal bodies, and cilia of drosophila melanogaster. Genetics 2017, 206, 33–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornens, M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002, 14, 25–34. [Google Scholar] [CrossRef]
- Ou, Y.Y.; Zhang, M.; Chi, S.; Matyas, J.R.; Rattner, J.B. Higher order structure of the pcm adjacent to the centriole. Cell Motil. Cytoskelet. 2003, 55, 125–133. [Google Scholar] [CrossRef]
- Rieder, C.L.; Jensen, C.G.; Jensen, L.C. The resorption of primary cilia during mitosis in a vertebrate (ptk1) cell line. J. Ultrastruct. Res. 1979, 68, 173–185. [Google Scholar] [CrossRef]
- De Brabander, M.; Geuens, G.; Nuydens, R.; Willebrords, R.; Aerts, F.; De Mey, J. Microtubule dynamics during the cell cycle: The effects of taxol and nocodazole on the microtubule system of pt k2 cells at different stages of the mitotic cycle. Int. Rev. Cytol. 1986, 101, 215–274. [Google Scholar]
- Kuriyama, R.; Borisy, G.G. Centriole cycle in chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 1981, 91, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, R.E.; Vogel, J.M.; Schnackenberg, B.J.; Hull, D.R.; Wu, X. Centrosome maturation. Curr. Top. Dev. Biol. 2000, 49, 449–470. [Google Scholar] [PubMed]
- O’Connell, K.F.; Caron, C.; Kopish, K.R.; Hurd, D.D.; Kemphues, K.J.; Li, Y.; White, J.G. The c. Elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 2001, 105, 547–558. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, K.F. The zyg-1 kinase, a mitotic and meiotic regulator of centriole replication. Oncogene 2002, 21, 6201–6208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemp, C.A.; Kopish, K.R.; Zipperlen, P.; Ahringer, J.; O’Connell, K.F. Centrosome maturation and duplication in c. Elegans require the coiled-coil protein spd-2. Dev. Cell 2004, 6, 511–523. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, L.; Ozlu, N.; Hannak, E.; Cowan, C.; Habermann, B.; Ruer, M.; Muller-Reichert, T.; Hyman, A.A. The caenorhabditis elegans centrosomal protein spd-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 2004, 14, 863–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkham, M.; Muller-Reichert, T.; Oegema, K.; Grill, S.; Hyman, A.A. Sas-4 is a c. Elegans centriolar protein that controls centrosome size. Cell 2003, 112, 575–587. [Google Scholar] [CrossRef] [Green Version]
- Leidel, S.; Gonczy, P. Sas-4 is essential for centrosome duplication in c elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 2003, 4, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Delattre, M.; Leidel, S.; Wani, K.; Baumer, K.; Bamat, J.; Schnabel, H.; Feichtinger, R.; Schnabel, R.; Gonczy, P. Centriolar sas-5 is required for centrosome duplication in c. Elegans. Nat. Cell Biol. 2004, 6, 656–664. [Google Scholar] [CrossRef]
- Dammermann, A.; Muller-Reichert, T.; Pelletier, L.; Habermann, B.; Desai, A.; Oegema, K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 2004, 7, 815–829. [Google Scholar] [CrossRef] [Green Version]
- Leidel, S.; Delattre, M.; Cerutti, L.; Baumer, K.; Gonczy, P. Sas-6 defines a protein family required for centrosome duplication in c. Elegans and in human cells. Nat. Cell Biol. 2005, 7, 115–125. [Google Scholar] [CrossRef]
- Habedanck, R.; Stierhof, Y.D.; Wilkinson, C.J.; Nigg, E.A. The polo kinase plk4 functions in centriole duplication. Nat. Cell Biol. 2005, 7, 1140–1146. [Google Scholar] [CrossRef]
- Bettencourt-Dias, M.; Rodrigues-Martins, A.; Carpenter, L.; Riparbelli, M.; Lehmann, L.; Gatt, M.K.; Carmo, N.; Balloux, F.; Callaini, G.; Glover, D.M. Sak/plk4 is required for centriole duplication and flagella development. Curr. Biol. 2005, 15, 2199–2207. [Google Scholar] [CrossRef] [Green Version]
- Strnad, P.; Gonczy, P. Mechanisms of procentriole formation. Trends Cell Biol. 2008, 18, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Blachon, S.; Gopalakrishnan, J.; Omori, Y.; Polyanovsky, A.; Church, A.; Nicastro, D.; Malicki, J.; Avidor-Reiss, T. Drosophila asterless and vertebrate cep152 are orthologs essential for centriole duplication. Genetics 2008, 180, 2081–2094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guernsey, D.L.; Jiang, H.; Hussin, J.; Arnold, M.; Bouyakdan, K.; Perry, S.; Babineau-Sturk, T.; Beis, J.; Dumas, N.; Evans, S.C.; et al. Mutations in centrosomal protein cep152 in primary microcephaly families linked to mcph4. Am. J. Hum. Genet. 2010, 87, 40–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatch, E.; Stearns, T. The life cycle of centrioles. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 425–431. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.J.; Marjanovic, M.; Luders, J.; Stracker, T.H.; Costanzo, V. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS ONE 2013, 8, e69986. [Google Scholar] [CrossRef] [Green Version]
- Lukinavicius, G.; Lavogina, D.; Orpinell, M.; Umezawa, K.; Reymond, L.; Garin, N.; Gonczy, P.; Johnsson, K. Selective chemical crosslinking reveals a cep57-cep63-cep152 centrosomal complex. Curr. Biol. 2013, 23, 265–270. [Google Scholar] [CrossRef] [Green Version]
- Rattner, J.B.; Phillips, S.G. Independence of centriole formation and DNA synthesis. J. Cell Biol. 1973, 57, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Sluder, G. Centrosomes and the cell cycle. J. Cell Sci. Suppl. 1989, 12, 253–275. [Google Scholar] [CrossRef] [Green Version]
- Gorgidze, L.A.; Vorobjev, I.A. Centrosome and microtubules behavior in the cytoplasts. J. Submicrosc. Cytol. Pathol. 1995, 27, 381–389. [Google Scholar]
- Uzbekov, R.E. Centriole duplication in PE (SPEV) cells starts before beginning of DNA replication. Biochem. Moscow Suppl. Ser. A 2007, 1, 206–211. [Google Scholar] [CrossRef]
- Uzbekov, R.E.; Alieva, I.B. Centriole duplication or DNA replication—What starts earlier? In Cytoskeleton: Cell Movement, Cytokinesis and Organelles Organization; Lansing, S., Rousseau, T., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2010; pp. 127–137. [Google Scholar]
- Hinchcliffe, E.H.; Li, C.; Thompson, E.A.; Maller, J.L.; Sluder, G. Requirement of cdk2-cyclin e activity for repeated centrosome reproduction in xenopus egg extracts. Science 1999, 283, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Lacey, K.R.; Jackson, P.K.; Stearns, T. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA 1999, 96, 2817–2822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meraldi, P.; Lukas, J.; Fry, A.M.; Bartek, J.; Nigg, E.A. Centrosome duplication in mammalian somatic cells requires e2f and cdk2-cyclin a. Nat. Cell Biol. 1999, 1, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Dirksen, E.R. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J. Cell Biol. 1971, 51, 286–302. [Google Scholar] [CrossRef] [Green Version]
- Nanjundappa, R.; Kong, D.; Shim, K.; Stearns, T.; Brody, S.L.; Loncarek, J.; Mahjoub, M.R. Regulation of cilia abundance in multiciliated cells. ELife 2019, 8, e44039. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Q.; Fang, C.; Huang, Q.; Zhou, J.; Yan, X.; Zhu, X. Parental centrioles are dispensable for deuterosome formation and function during basal body amplification. EMBO Rep. 2019, 20, e46735. [Google Scholar] [CrossRef]
- Szollosi, D.; Calarco, P.; Donahue, R.P. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 1972, 11, 521–541. [Google Scholar]
- Khodjakov, A.; Rieder, C.L.; Sluder, G.; Cassels, G.; Sibon, O.; Wang, C.L. De novo formation of centrosomes in vertebrate cells arrested during s phase. J. Cell Biol. 2002, 158, 1171–1181. [Google Scholar] [CrossRef] [Green Version]
- Callaini, G.; Riparbelli, M.G.; Dallai, R. Centrosome inheritance in insects: Fertilization and parthenogenesis. Biol. Cell 1999, 91, 355–366. [Google Scholar] [CrossRef]
- Fawcett, D.W.; Phillips, D.M. The fine structure and development of the neck region of the mammalian spermatozoon. Anat. Rec. 1969, 165, 153–164. [Google Scholar] [CrossRef]
- Garanina, A.S.; Alieva, I.B.; Bragina, E.E.; Blanchard, E.; Arbeille, B.; Guerif, F.; Uzbekova, S.; Uzbekov, R.E. The centriolar adjunct-appearance and disassembly in spermiogenesis and the potential impact on fertility. Cells 2019, 8, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alieva, I.B.; Staub, C.; Uzbekova, S.; Uzbekov, R.E. A question of flagella origin for spermatids—Mother or daughter centriole? In Flagella and Cilia. Types Structure and Functions; Uzbekov, R.E., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2018; pp. 109–126. [Google Scholar]
- Sullenberger, C.; Vasquez-Limeta, A.; Kong, D.; Loncarek, J. With age comes maturity: Biochemical and structural transformation of a human centriole in the making. Cells 2020, 9, 1429. [Google Scholar] [CrossRef] [PubMed]
- Robbins, E.; Jentzsch, G.; Micali, A. The centriole cycle in synchronized hela cells. J. Cell Biol. 1968, 36, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sluder, G. Using sea urchin gametes and zygotes to investigate centrosome duplication. Cilia 2016, 5, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigg, E.A.; Stearns, T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011, 13, 1154–1160. [Google Scholar] [CrossRef] [Green Version]
- Izquierdo, D.; Wang, W.J.; Uryu, K.; Tsou, M.F. Stabilization of cartwheel-less centrioles for duplication requires cep295-mediated centriole-to-centrosome conversion. Cell Rep. 2014, 8, 957–965. [Google Scholar] [CrossRef] [Green Version]
- Nigg, E.A.; Holland, A.J. Once and only once: Mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 297–312. [Google Scholar] [CrossRef]
- Inoue, D.; Obino, D.; Pineau, J.; Farina, F.; Gaillard, J.; Guerin, C.; Blanchoin, L.; Lennon-Dumenil, A.M.; Thery, M. Actin filaments regulate microtubule growth at the centrosome. EMBO J. 2019, 38, e99630. [Google Scholar] [CrossRef]
- Dujardin, D.L.; Vallee, R.B. Dynein at the cortex. Curr. Opin. Cell Biol. 2002, 14, 44–49. [Google Scholar] [CrossRef]
- Inoue, S.; Salmon, E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 1995, 6, 1619–1640. [Google Scholar] [CrossRef]
- Holy, T.E.; Dogterom, M.; Yurke, B.; Leibler, S. Assembly and positioning of microtubule asters in microfabricated chambers. Proc. Natl. Acad. Sci. USA 1997, 94, 6228–6231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmon, W.C.; Adams, M.C.; Waterman-Storer, C.M. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 2002, 158, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Burakov, A.; Nadezhdina, E.; Slepchenko, B.; Rodionov, V. Centrosome positioning in interphase cells. J. Cell Biol. 2003, 162, 963–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burakov, A.V.; Nadezhdina, E.S. Centering and shifting of centrosomes in cells. Cells 2020, 9, 1351. [Google Scholar] [CrossRef] [PubMed]
- Goldman, R.D. The role of three cytoplasmic fibers in bhk-21 cell motility. I. Microtubules and the effects of colchicine. J. Cell Biol. 1971, 51, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alieva, I.B.; Vaisberg, E.A.; Nadezhdina, E.S.; Vorobjev, I.A. Microtubule and intermediate filament patterns around the centrosome in interphase cells. In The Centrosome; Kalnins, V.C., Ed.; Academic Press: Cambridge, MA, USA, 1991; pp. 103–129. [Google Scholar]
- Avidor-Reiss, T. Rapid evolution of sperm produces diverse centriole structures that reveal the most rudimentary structure needed for function. Cells 2018, 7, 67. [Google Scholar] [CrossRef] [Green Version]
- Avidor-Reiss, T.; Fishman, E.L. It takes two (centrioles) to tango. Reproduction 2019, 157, R33–R51. [Google Scholar] [CrossRef] [Green Version]
- Khire, A.; Jo, K.H.; Kong, D.; Akhshi, T.; Blachon, S.; Cekic, A.R.; Hynek, S.; Ha, A.; Loncarek, J.; Mennella, V.; et al. Centriole remodeling during spermiogenesis in drosophila. Curr. Biol. 2016, 26, 3183–3189. [Google Scholar] [CrossRef] [Green Version]
- Fishman, E.L.; Jo, K.; Ha, A.; Royfman, R.; Zinn, A.; Krishnamurthy, M.; Avidor-Reiss, T. Atypical centrioles are present in tribolium sperm. Open Biol. 2017, 7, 160334. [Google Scholar] [CrossRef] [Green Version]
- Huettner, A.F.; Rabinowitz, M. Demonstration of the central body in the living cell. Science 1933, 78, 367–368. [Google Scholar] [CrossRef]
- Tassin, A.M.; Maro, B.; Bornens, M. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 1985, 100, 35–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manandhar, G.; Sutovsky, P.; Joshi, H.C.; Stearns, T.; Schatten, G. Centrosome reduction during mouse spermiogenesis. Dev. Biol. 1998, 203, 424–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulton, C.; Dingle, A.D. Basal bodies, but not centrioles, in naegleria. J. Cell Biol. 1971, 51, 826–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz-Laylin, L.K.; Fulton, C. Naegleria: A classic model for de novo basal body assembly. Cilia 2016, 5, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aprelikova, O.N.; Fang, B.S.; Meissner, E.G.; Cotter, S.; Campbell, M.; Kuthiala, A.; Bessho, M.; Jensen, R.A.; Liu, E.T. Brca1-associated growth arrest is rb-dependent. Proc. Natl. Acad. Sci. USA 1999, 96, 11866–11871. [Google Scholar] [CrossRef] [Green Version]
- Arquint, C.; Nigg, E.A. The plk4-stil-sas-6 module at the core of centriole duplication. Biochem. Soc. Trans. 2016, 44, 1253–1263. [Google Scholar] [CrossRef] [Green Version]
- Askham, J.M.; Vaughan, K.T.; Goodson, H.V.; Morrison, E.E. Evidence that an interaction between eb1 and p150(glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol. Biol. Cell 2002, 13, 3627–3645. [Google Scholar] [CrossRef] [Green Version]
- Astuti, P.; Boutros, R.; Ducommun, B.; Gabrielli, B. Mitotic phosphorylation of cdc25b ser321 disrupts 14-3-3 binding to the high affinity ser323 site. J. Biol. Chem. 2010, 285, 34364–34370. [Google Scholar] [CrossRef] [Green Version]
- Atorino, E.S.; Hata, S.; Funaya, C.; Neuner, A.; Schiebel, E. Cep44 ensures the formation of bona fide centriole wall, a requirement for the centriole-to-centrosome conversion. Nat. Commun. 2020, 11, 903. [Google Scholar] [CrossRef]
- Barrera, J.A.; Kao, L.R.; Hammer, R.E.; Seemann, J.; Fuchs, J.L.; Megraw, T.L. Cdk5rap2 regulates centriole engagement and cohesion in mice. Dev. Cell 2010, 18, 913–926. [Google Scholar] [CrossRef] [Green Version]
- Basto, R.; Lau, J.; Vinogradova, T.; Gardiol, A.; Woods, C.G.; Khodjakov, A.; Raff, J.W. Flies without centrioles. Cell 2006, 125, 1375–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastos, R.N.; Barr, F.A. Plk1 negatively regulates cep55 recruitment to the midbody to ensure orderly abscission. J. Cell Biol. 2010, 191, 751–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrueta, L.; Kraeft, S.K.; Tirnauer, J.S.; Schuyler, S.C.; Chen, L.B.; Hill, D.E.; Pellman, D.; Bierer, B.E. The adenomatous polyposis coli-binding protein eb1 is associated with cytoplasmic and spindle microtubules. Proc. Natl. Acad. Sci. USA 1998, 95, 10596–10601. [Google Scholar] [CrossRef] [Green Version]
- Bowler, M.; Kong, D.; Sun, S.; Nanjundappa, R.; Evans, L.; Farmer, V.; Holland, A.; Mahjoub, M.R.; Sui, H.; Loncarek, J. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative storm and electron microscopy. Nat. Commun. 2019, 10, 993. [Google Scholar] [CrossRef]
- Brown, C.R.; Doxsey, S.J.; Hong-Brown, L.Q.; Martin, R.L.; Welch, W.J. Molecular chaperones and the centrosome. A role for tcp-1 in microtubule nucleation. J. Biol. Chem. 1996, 271, 824–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.R.; Hong-Brown, L.Q.; Doxsey, S.J.; Welch, W.J. Molecular chaperones and the centrosome. A role for hsp 73 in centrosomal repair following heat shock treatment. J. Biol. Chem. 1996, 271, 833–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brueckner, M. Cilia propel the embryo in the right direction. Am. J. Med Genet. 2001, 101, 339–344. [Google Scholar] [CrossRef]
- Burakov, A.; Kovalenko, O.; Semenova, I.; Zhapparova, O.; Nadezhdina, E.; Rodionov, V. Cytoplasmic dynein is involved in the retention of microtubules at the centrosome in interphase cells. Traffic 2008, 9, 472–480. [Google Scholar] [CrossRef]
- Cajanek, L.; Nigg, E.A. Cep164 triggers ciliogenesis by recruiting tau tubulin kinase 2 to the mother centriole. Proc. Natl. Acad. Sci. USA 2014, 111, E2841–E2850. [Google Scholar] [CrossRef] [Green Version]
- Carroll, P.E.; Okuda, M.; Horn, H.F.; Biddinger, P.; Stambrook, P.J.; Gleich, L.L.; Li, Y.Q.; Tarapore, P.; Fukasawa, K. Centrosome hyperamplification in human cancer: Chromosome instability induced by p53 mutation and/or mdm2 overexpression. Oncogene 1999, 18, 1935–1944. [Google Scholar] [CrossRef] [Green Version]
- Casenghi, M.; Meraldi, P.; Weinhart, U.; Duncan, P.I.; Korner, R.; Nigg, E.A. Polo-like kinase 1 regulates nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 2003, 5, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Casenghi, M.; Barr, F.A.; Nigg, E.A. Phosphorylation of nlp by plk1 negatively regulates its dynein-dynactin-dependent targeting to the centrosome. J. Cell Sci. 2005, 118, 5101–5108. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.; Stearns, T. Delta-tubulin and epsilon-tubulin: Two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2000, 2, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Cizmecioglu, O.; Hoffmann, I.; Rhee, K. Plk2 phosphorylation is critical for cpap function in procentriole formation during the centrosome cycle. EMBO J. 2010, 29, 2395–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chartier, N.T.; Oddou, C.I.; Laine, M.G.; Ducarouge, B.; Marie, C.A.; Block, M.R.; Jacquier-Sarlin, M.R. Cyclin-dependent kinase 2/cyclin e complex is involved in p120 catenin (p120ctn)-dependent cell growth control: A new role for p120ctn in cancer. Cancer Res. 2007, 67, 9781–9790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Indjeian, V.B.; McManus, M.; Wang, L.; Dynlacht, B.D. Cp110, a cell cycle-dependent cdk substrate, regulates centrosome duplication in human cells. Dev. Cell 2002, 3, 339–350. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Howng, S.L.; Cheng, T.S.; Chou, M.H.; Huang, C.Y.; Hong, Y.R. Molecular characterization of human ninein protein: Two distinct subdomains required for centrosomal targeting and regulating signals in cell cycle. Biochem. Biophys. Res. Commun. 2003, 308, 975–983. [Google Scholar] [CrossRef]
- Chen, C.Y.; Olayioye, M.A.; Lindeman, G.J.; Tang, T.K. Cpap interacts with 14-3-3 in a cell cycle-dependent manner. Biochem. Biophys. Res. Commun. 2006, 342, 1203–1210. [Google Scholar] [CrossRef]
- Chen, H.Y.; Wu, C.T.; Tang, C.C.; Lin, Y.N.; Wang, W.J.; Tang, T.K. Human microcephaly protein rttn interacts with stil and is required to build full-length centrioles. Nat. Commun. 2017, 8, 247. [Google Scholar] [CrossRef]
- Cheng, T.S.; Hsiao, Y.L.; Lin, C.C.; Hsu, C.M.; Chang, M.S.; Lee, C.I.; Yu, R.C.; Huang, C.Y.; Howng, S.L.; Hong, Y.R. Hninein is required for targeting spindle-associated protein astrin to the centrosome during the s and g2 phases. Exp. Cell Res. 2007, 313, 1710–1721. [Google Scholar] [CrossRef]
- Choi, Y.K.; Liu, P.; Sze, S.K.; Dai, C.; Qi, R.Z. Cdk5rap2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 2010, 191, 1089–1095. [Google Scholar] [CrossRef] [Green Version]
- Cogswell, J.P.; Brown, C.E.; Bisi, J.E.; Neill, S.D. Dominant-negative polo-like kinase 1 induces mitotic catastrophe independent of cdc25c function. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 2000, 11, 615–623. [Google Scholar]
- Collins, E.S.; Hornick, J.E.; Durcan, T.M.; Collins, N.S.; Archer, W.; Karanjeet, K.B.; Vaughan, K.T.; Hinchcliffe, E.H. Centrosome biogenesis continues in the absence of microtubules during prolonged s-phase arrest. J. Cell. Physiol. 2010, 225, 454–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, E.; Marine, J.C.; Danovi, D.; Falini, B.; Pelicci, P.G. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002, 4, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Conduit, P.T.; Brunk, K.; Dobbelaere, J.; Dix, C.I.; Lucas, E.P.; Raff, J.W. Centrioles regulate centrosome size by controlling the rate of cnn incorporation into the pcm. Curr. Biol. 2010, 20, 2178–2186. [Google Scholar] [CrossRef] [Green Version]
- D’Angiolella, V.; Donato, V.; Vijayakumar, S.; Saraf, A.; Florens, L.; Washburn, M.P.; Dynlacht, B.; Pagano, M. Scf(cyclin f) controls centrosome homeostasis and mitotic fidelity through cp110 degradation. Nature 2010, 466, 138–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delattre, M.; Canard, C.; Gonczy, P. Sequential protein recruitment in c. Elegans centriole formation. Curr. Biol. 2006, 16, 1844–1849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diviani, D.; Langeberg, L.K.; Doxsey, S.J.; Scott, J.D. Pericentrin anchors protein kinase a at the centrosome through a newly identified rii-binding domain. Curr. Biol. 2000, 10, 417–420. [Google Scholar] [CrossRef] [Green Version]
- Doxsey, S.J.; Stein, P.; Evans, L.; Calarco, P.D.; Kirschner, M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 1994, 76, 639–650. [Google Scholar] [CrossRef]
- El Din El Homasany, B.S.; Volkov, Y.; Takahashi, M.; Ono, Y.; Keryer, G.; Delouvee, A.; Looby, E.; Long, A.; Kelleher, D. The scaffolding protein cg-nap/akap450 is a critical integrating component of the lfa-1-induced signaling complex in migratory t cells. J. Immunol. 2005, 175, 7811–7818. [Google Scholar] [CrossRef]
- Ellinger-Ziegelbauer, H.; Karasuyama, H.; Yamada, E.; Tsujikawa, K.; Todokoro, K.; Nishida, E. Ste20-like kinase (slk), a regulatory kinase for polo-like kinase (plk) during the g2/m transition in somatic cells. Genes Cells 2000, 5, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Eyers, P.A.; Erikson, E.; Chen, L.G.; Maller, J.L. A novel mechanism for activation of the protein kinase aurora a. Curr. Biol. 2003, 13, 691–697. [Google Scholar] [CrossRef]
- Fabbro, M.; Zhou, B.B.; Takahashi, M.; Sarcevic, B.; Lal, P.; Graham, M.E.; Gabrielli, B.G.; Robinson, P.J.; Nigg, E.A.; Ono, Y.; et al. Cdk1/erk2- and plk1-dependent phosphorylation of a centrosome protein, cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell 2005, 9, 477–488. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Yuan, J.H.; Maloid, S.C.; Fisher, R.; Copeland, T.D.; Longo, D.L.; Conrads, T.P.; Veenstra, T.D.; Ferris, A.; Hughes, S.; et al. Polo-like kinase 1-mediated phosphorylation of the gtp-binding protein ran is important for bipolar spindle formation. Biochem. Biophys. Res. Commun. 2006, 349, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Fong, K.W.; Choi, Y.K.; Rattner, J.B.; Qi, R.Z. Cdk5rap2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 2008, 19, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, C.S.; Ozaki, K.; Tsou, M.B. Ppp1r35 ensures centriole homeostasis by promoting centriole-to-centrosome conversion. Mol. Biol. Cell 2018, 29, 2801–2808. [Google Scholar] [CrossRef]
- Franck, N.; Montembault, E.; Rome, P.; Pascal, A.; Cremet, J.Y.; Giet, R. Cdk11(p58) is required for centriole duplication and plk4 recruitment to mitotic centrosomes. PLoS ONE 2011, 6, e14600. [Google Scholar] [CrossRef] [Green Version]
- Frank-Vaillant, M.; Haccard, O.; Thibier, C.; Ozon, R.; Arlot-Bonnemains, Y.; Prigent, C.; Jessus, C. Progesterone regulates the accumulation and the activation of eg2 kinase in xenopus oocytes. J. Cell Sci. 2000, 113, 1127–1138. [Google Scholar]
- Fry, A.M.; Mayor, T.; Meraldi, P.; Stierhof, Y.D.; Tanaka, K.; Nigg, E.A. C-nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase nek2. J. Cell Biol. 1998, 141, 1563–1574. [Google Scholar] [CrossRef] [Green Version]
- Fry, A.M.; Meraldi, P.; Nigg, E.A. A centrosomal function for the human nek2 protein kinase, a member of the nima family of cell cycle regulators. EMBO J. 1998, 17, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Hagan, I.M.; Glover, D.M. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 2015, 7, a015800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukasawa, K. P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim. Biophys. Acta 2008, 1786, 15–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giet, R.; Uzbekov, R.; Kireev, I.; Prigent, C. The xenopus laevis centrosome aurora/ipl1-related kinase. Biol. Cell 1999, 91, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Gluenz, E.; Hoog, J.L.; Smith, A.E.; Dawe, H.R.; Shaw, M.K.; Gull, K. Beyond 9+0: Noncanonical axoneme structures characterize sensory cilia from protists to humans. FASEB J. 2010, 24, 3117–3121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopalakrishnan, J.; Mennella, V.; Blachon, S.; Zhai, B.; Smith, A.H.; Megraw, T.L.; Nicastro, D.; Gygi, S.P.; Agard, D.A.; Avidor-Reiss, T. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2011, 2, 359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, M.; O’Brien, D.A.; Eddy, E.M. Speriolin is a novel human and mouse sperm centrosome protein. Hum. Reprod. 2010, 25, 1884–1894. [Google Scholar] [CrossRef] [Green Version]
- Graser, S.; Stierhof, Y.D.; Lavoie, S.B.; Gassner, O.S.; Lamla, S.; Le Clech, M.; Nigg, E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007, 179, 321–330. [Google Scholar] [CrossRef] [Green Version]
- Groen, A.C.; Cameron, L.A.; Coughlin, M.; Miyamoto, D.T.; Mitchison, T.J.; Ohi, R. Xrhamm functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 2004, 14, 1801–1811. [Google Scholar] [CrossRef] [Green Version]
- Gromley, A.; Yeaman, C.; Rosa, J.; Redick, S.; Chen, C.T.; Mirabelle, S.; Guha, M.; Sillibourne, J.; Doxsey, S.J. Centriolin anchoring of exocyst and snare complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 2005, 123, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Guarguaglini, G.; Duncan, P.I.; Stierhof, Y.D.; Holmstrom, T.; Duensing, S.; Nigg, E.A. The forkhead-associated domain protein cep170 interacts with polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell 2005, 16, 1095–1107. [Google Scholar] [CrossRef] [Green Version]
- Gudi, R.; Zou, C.; Li, J.; Gao, Q. Centrobin-tubulin interaction is required for centriole elongation and stability. J. Cell Biol. 2011, 193, 711–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillet, V.; Knibiehler, M.; Gregory-Pauron, L.; Remy, M.H.; Chemin, C.; Raynaud-Messina, B.; Bon, C.; Kollman, J.M.; Agard, D.A.; Merdes, A.; et al. Crystal structure of gamma-tubulin complex protein gcp4 provides insight into microtubule nucleation. Nat. Struct. Mol. Biol. 2011, 18, 915–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Yang, Z.; Song, W.; Chen, Q.; Wang, F.; Zhang, Q.; Zhu, X. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell 2006, 17, 680–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurkaslar, H.K.; Culfa, E.; Arslanhan, M.D.; Lince-Faria, M.; Firat-Karalar, E.N. Ccdc57 cooperates with microtubules and microcephaly protein cep63 and regulates centriole duplication and mitotic progression. Cell Rep. 2020, 31, 107630. [Google Scholar] [CrossRef]
- Hamel, V.; Steib, E.; Hamelin, R.; Armand, F.; Borgers, S.; Fluckiger, I.; Busso, C.; Olieric, N.; Sorzano, C.O.S.; Steinmetz, M.O.; et al. Identification of chlamydomonas central core centriolar proteins reveals a role for human wdr90 in ciliogenesis. Curr. Biol. 2017, 27, 2486–2498.e6. [Google Scholar] [CrossRef] [Green Version]
- Hames, R.S.; Crookes, R.E.; Straatman, K.R.; Merdes, A.; Hayes, M.J.; Faragher, A.J.; Fry, A.M. Dynamic recruitment of nek2 kinase to the centrosome involves microtubules, pcm-1, and localized proteasomal degradation. Mol. Biol. Cell 2005, 16, 1711–1724. [Google Scholar] [CrossRef]
- Harbour, J.W.; Luo, R.X.; Dei Santi, A.; Postigo, A.A.; Dean, D.C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block rb functions as cells move through g1. Cell 1999, 98, 859–869. [Google Scholar] [CrossRef] [Green Version]
- Haren, L.; Stearns, T.; Luders, J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple pcm components. PLoS ONE 2009, 4, e5976. [Google Scholar] [CrossRef]
- Hirayama, S.; Yamazaki, Y.; Kitamura, A.; Oda, Y.; Morito, D.; Okawa, K.; Kimura, H.; Cyr, D.M.; Kubota, H.; Nagata, K. Mkks is a centrosome-shuttling protein degraded by disease-causing mutations via chip-mediated ubiquitination. Mol. Biol. Cell 2008, 19, 899–911. [Google Scholar] [CrossRef] [Green Version]
- Hirota, T.; Kunitoku, N.; Sasayama, T.; Marumoto, T.; Zhang, D.; Nitta, M.; Hatakeyama, K.; Saya, H. Aurora-a and an interacting activator, the lim protein ajuba, are required for mitotic commitment in human cells. Cell 2003, 114, 585–598. [Google Scholar] [CrossRef] [Green Version]
- Hosono, K.; Noda, S.; Shimizu, A.; Nakanishi, N.; Ohtsubo, M.; Shimizu, N.; Minoshima, S. Ypel5 protein of the ypel gene family is involved in the cell cycle progression by interacting with two distinct proteins ranbpm and ranbp10. Genomics 2010, 96, 102–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, L.C.; White, R.L. Brca1 is associated with the centrosome during mitosis. Proc. Natl. Acad. Sci. USA 1998, 95, 12983–12988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Xia, Y.; Zhang, D.; Wang, S.; Bao, Y.; He, R.; Teng, J.; Chen, J. Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins ccdc120 and ccdc68. Nat. Commun. 2017, 8, 15057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.H.; Wu, C.C.; Chou, C.K.; Huang, C.Y. A translational regulator, pum2, promotes both protein stability and kinase activity of aurora-a. PLoS ONE 2011, 6, e19718. [Google Scholar] [CrossRef] [Green Version]
- Ibi, M.; Zou, P.; Inoko, A.; Shiromizu, T.; Matsuyama, M.; Hayashi, Y.; Enomoto, M.; Mori, D.; Hirotsune, S.; Kiyono, T.; et al. Trichoplein controls microtubule anchoring at the centrosome by binding to odf2 and ninein. J. Cell Sci. 2011, 124, 857–864. [Google Scholar] [CrossRef] [Green Version]
- Iwamori, T.; Iwamori, N.; Ma, L.; Edson, M.A.; Greenbaum, M.P.; Matzuk, M.M. Tex14 interacts with cep55 to block cell abscission. Mol. Cell. Biol. 2010, 30, 2280–2292. [Google Scholar] [CrossRef] [Green Version]
- Jackman, M.; Lindon, C.; Nigg, E.A.; Pines, J. Active cyclin b1-cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003, 5, 143–148. [Google Scholar] [CrossRef]
- Jakobsen, L.; Vanselow, K.; Skogs, M.; Toyoda, Y.; Lundberg, E.; Poser, I.; Falkenby, L.G.; Bennetzen, M.; Westendorf, J.; Nigg, E.A.; et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011, 30, 1520–1535. [Google Scholar] [CrossRef]
- Jang, Y.J.; Ji, J.H.; Ahn, J.H.; Hoe, K.L.; Won, M.; Im, D.S.; Chae, S.K.; Song, S.; Yoo, H.S. Polo-box motif targets a centrosome regulator, rangtpase. Biochem. Biophys. Res. Commun. 2004, 325, 257–264. [Google Scholar] [CrossRef]
- Jin, S.; Gao, H.; Mazzacurati, L.; Wang, Y.; Fan, W.; Chen, Q.; Yu, W.; Wang, M.; Zhu, X.; Zhang, C.; et al. Brca1 interaction of centrosomal protein nlp is required for successful mitotic progression. J. Biol. Chem. 2009, 284, 22970–22977. [Google Scholar] [CrossRef] [Green Version]
- Jo, K.H.; Jaiswal, A.; Khanal, S.; Fishman, E.L.; Curry, A.N.; Avidor-Reiss, T. Poc1b and sas-6 function together during the atypical centriole formation in drosophila melanogaster. Cells 2019, 8, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joukov, V.; Groen, A.C.; Prokhorova, T.; Gerson, R.; White, E.; Rodriguez, A.; Walter, J.C.; Livingston, D.M. The brca1/bard1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 2006, 127, 539–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, L.R.; Megraw, T.L. Centrocortin cooperates with centrosomin to organize drosophila embryonic cleavage furrows. Curr. Biol. 2009, 19, 937–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katayama, H.; Sasai, K.; Kawai, H.; Yuan, Z.M.; Bondaruk, J.; Suzuki, F.; Fujii, S.; Arlinghaus, R.B.; Czerniak, B.A.; Sen, S. Phosphorylation by aurora kinase a induces mdm2-mediated destabilization and inhibition of p53. Nat. Genet. 2004, 36, 55–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellogg, D.R.; Moritz, M.; Alberts, B.M. The centrosome and cellular organization. Annu. Rev. Biochem. 1994, 63, 639–674. [Google Scholar] [CrossRef]
- Keryer, G.; Di Fiore, B.; Celati, C.; Lechtreck, K.F.; Mogensen, M.; Delouvee, A.; Lavia, P.; Bornens, M.; Tassin, A.M. Part of ran is associated with akap450 at the centrosome: Involvement in microtubule-organizing activity. Mol. Biol. Cell 2003, 14, 4260–4271. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Takahashi, M.; Matsuo, K.; Ono, Y. Recruitment of cg-nap to the golgi apparatus through interaction with dynein-dynactin complex. Genes Cells 2007, 12, 421–434. [Google Scholar] [CrossRef]
- Kim, J.C.; Ou, Y.Y.; Badano, J.L.; Esmail, M.A.; Leitch, C.C.; Fiedrich, E.; Beales, P.L.; Archibald, J.M.; Katsanis, N.; Rattner, J.B.; et al. Mkks/bbs6, a divergent chaperonin-like protein linked to the obesity disorder bardet-biedl syndrome, is a novel centrosomal component required for cytokinesis. J. Cell Sci. 2005, 118, 1007–1020. [Google Scholar] [CrossRef] [Green Version]
- Kleylein-Sohn, J.; Westendorf, J.; Le Clech, M.; Habedanck, R.; Stierhof, Y.D.; Nigg, E.A. Plk4-induced centriole biogenesis in human cells. Dev. Cell 2007, 13, 190–202. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Kim, S.; Lin, Y.C.; Inoue, T.; Dynlacht, B.D. The cp110-interacting proteins talpid3 and cep290 play overlapping and distinct roles in cilia assembly. J. Cell Biol. 2014, 204, 215–229. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Tsang, W.Y.; Li, J.; Lane, W.; Dynlacht, B.D. Centriolar kinesin kif24 interacts with cp110 to remodel microtubules and regulate ciliogenesis. Cell 2011, 145, 914–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotani, T.; Yamashita, M. Overexpression of truncated gamma-tubulins disrupts mitotic aster formation in xenopus oocyte extracts. Biochem. J. 2005, 389, 611–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, A.; Hoffmann, I. Polo-like kinase 2-dependent phosphorylation of npm/b23 on serine 4 triggers centriole duplication. PLoS ONE 2010, 5, e9849. [Google Scholar] [CrossRef] [Green Version]
- Kurz, T.; Pintard, L.; Willis, J.H.; Hamill, D.R.; Gonczy, P.; Peter, M.; Bowerman, B. Cytoskeletal regulation by the nedd8 ubiquitin-like protein modification pathway. Science 2002, 295, 1294–1298. [Google Scholar] [CrossRef]
- Lee, M.J.; Gergely, F.; Jeffers, K.; Peak-Chew, S.Y.; Raff, J.W. Msps/xmap215 interacts with the centrosomal protein d-tacc to regulate microtubule behaviour. Nat. Cell Biol. 2001, 3, 643–649. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Rhee, K. Cep215 is involved in the dynein-dependent accumulation of pericentriolar matrix proteins for spindle pole formation. Cell Cycle 2010, 9, 774–783. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, J.; Jiao, H.; Liao, J.; Xu, X. Cytokinesis and cancer: Polo loves rock’n’ rho(a). J. Genet. Genom. = Yi Chuan Xue Bao 2010, 37, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhan, Q. The role of centrosomal nlp in the control of mitotic progression and tumourigenesis. Br. J. Cancer 2011, 104, 1523–1528. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Hansen, D.; Killilea, A.; Joshi, H.C.; Palazzo, R.E.; Balczon, R. Kendrin/pericentrin-b, a centrosome protein with homology to pericentrin that complexes with pcm-1. J. Cell Sci. 2001, 114, 797–809. [Google Scholar]
- Lin, C.C.; Cheng, T.S.; Hsu, C.M.; Wu, C.H.; Chang, L.S.; Shen, Z.S.; Yeh, H.M.; Chang, L.K.; Howng, S.L.; Hong, Y.R. Characterization and functional aspects of human ninein isoforms that regulated by centrosomal targeting signals and evidence for docking sites to direct gamma-tubulin. Cell Cycle 2006, 5, 2517–2527. [Google Scholar] [CrossRef] [Green Version]
- Linck, R.W.; Albertini, D.F.; Kenney, D.M.; Langevin, G.L. Tektin filaments: Chemically unique filaments of sperm flagellar microtubules. Prog. Clin. Biol. Res. 1982, 80, 127–132. [Google Scholar] [CrossRef]
- Lou, Y.; Xie, W.; Zhang, D.F.; Yao, J.H.; Luo, Z.F.; Wang, Y.Z.; Shi, Y.Y.; Yao, X.B. Nek2a specifies the centrosomal localization of erk2. Biochem. Biophys. Res. Commun. 2004, 321, 495–501. [Google Scholar] [CrossRef]
- Ma, S.; Trivinos-Lagos, L.; Graf, R.; Chisholm, R.L. Dynein intermediate chain mediated dynein-dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in dictyostelium. J. Cell Biol. 1999, 147, 1261–1274. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Kanai, M.; Kawamura, K.; Kaibuchi, K.; Ye, K.; Fukasawa, K. Interaction between rock ii and nucleophosmin/b23 in the regulation of centrosome duplication. Mol. Cell. Biol. 2006, 26, 9016–9034. [Google Scholar] [CrossRef] [Green Version]
- Mardin, B.R.; Lange, C.; Baxter, J.E.; Hardy, T.; Scholz, S.R.; Fry, A.M.; Schiebel, E. Components of the hippo pathway cooperate with nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 2010, 12, 1166–1176. [Google Scholar] [CrossRef] [Green Version]
- Mariappan, A.; Soni, K.; Schorpp, K.; Zhao, F.; Minakar, A.; Zheng, X.; Mandad, S.; Macheleidt, I.; Ramani, A.; Kubelka, T.; et al. Inhibition of cpap-tubulin interaction prevents proliferation of centrosome-amplified cancer cells. EMBO J. 2019, 38, e99876. [Google Scholar] [CrossRef]
- Matsuo, K.; Nishimura, T.; Hayakawa, A.; Ono, Y.; Takahashi, M. Involvement of a centrosomal protein kendrin in the maintenance of centrosome cohesion by modulating nek2a kinase activity. Biochem. Biophys. Res. Commun. 2010, 398, 217–223. [Google Scholar] [CrossRef]
- Maxwell, C.A.; Keats, J.J.; Belch, A.R.; Pilarski, L.M.; Reiman, T. Receptor for hyaluronan-mediated motility correlates with centrosome abnormalities in multiple myeloma and maintains mitotic integrity. Cancer Res. 2005, 65, 850–860. [Google Scholar]
- May-Simera, H.L.; Ross, A.; Rix, S.; Forge, A.; Beales, P.L.; Jagger, D.J. Patterns of expression of bardet-biedl syndrome proteins in the mammalian cochlea suggest noncentrosomal functions. J. Comp. Neurol. 2009, 514, 174–188. [Google Scholar] [CrossRef]
- McKendrick, L.; Milne, D.; Meek, D. Protein kinase ck2-dependent regulation of p53 function: Evidence that the phosphorylation status of the serine 386 (ck2) site of p53 is constitutive and stable. Mol. Cell. Biochem. 1999, 191, 187–199. [Google Scholar] [CrossRef]
- McNally, F.J.; Vale, R.D. Identification of katanin, an atpase that severs and disassembles stable microtubules. Cell 1993, 75, 419–429. [Google Scholar] [CrossRef]
- Megraw, T.L.; Li, K.; Kao, L.R.; Kaufman, T.C. The centrosomin protein is required for centrosome assembly and function during cleavage in drosophila. Development 1999, 126, 2829–2839. [Google Scholar]
- Merdes, A.; Cleveland, D.W. Pathways of spindle pole formation: Different mechanisms; conserved components. J. Cell Biol. 1997, 138, 953–956. [Google Scholar] [CrossRef] [Green Version]
- Merdes, A.; Ramyar, K.; Vechio, J.D.; Cleveland, D.W. A complex of numa and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 1996, 87, 447–458. [Google Scholar] [CrossRef] [Green Version]
- Mi, J.; Guo, C.; Brautigan, D.L.; Larner, J.M. Protein phosphatase-1alpha regulates centrosome splitting through nek2. Cancer Res. 2007, 67, 1082–1089. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, K.; Asanuma, M.; Miyazaki, I.; Diaz-Corrales, F.J.; Katayama, T.; Tohyama, M.; Ogawa, N. Disc1 localizes to the centrosome by binding to kendrin. Biochem. Biophys. Res. Commun. 2004, 317, 1195–1199. [Google Scholar] [CrossRef]
- Mogensen, M.M. Microtubule organizing centers in polarized epithelial cells. In Centrosomes in Development and Disease; Nigg, E.A., Ed.; Wiley-VCH Verlag GmbH & Co., KGaA: Weinheim, Germany, 2004. [Google Scholar]
- Mogensen, M.M.; Malik, A.; Piel, M.; Bouckson-Castaing, V.; Bornens, M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: The role of ninein. J. Cell Sci. 2000, 113, 3013–3023. [Google Scholar]
- Molli, P.R.; Li, D.Q.; Bagheri-Yarmand, R.; Pakala, S.B.; Katayama, H.; Sen, S.; Iyer, J.; Chernoff, J.; Tsai, M.Y.; Nair, S.S.; et al. Arpc1b, a centrosomal protein, is both an activator and substrate of aurora a. J. Cell Biol. 2010, 190, 101–114. [Google Scholar] [CrossRef]
- Morris, J.A.; Kandpal, G.; Ma, L.; Austin, C.P. Disc1 (disrupted-in-schizophrenia 1) is a centrosome-associated protein that interacts with map1a, mipt3, atf4/5 and nudel: Regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003, 12, 1591–1608. [Google Scholar] [CrossRef]
- Moudjou, M.; Bordes, N.; Paintrand, M.; Bornens, M. Gamma-tubulin in mammalian cells: The centrosomal and the cytosolic forms. J. Cell Sci. 1996, 109, 875–887. [Google Scholar]
- Munemitsu, S.; Souza, B.; Muller, O.; Albert, I.; Rubinfeld, B.; Polakis, P. The apc gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer Res. 1994, 54, 3676–3681. [Google Scholar] [PubMed]
- Murphy, S.M.; Preble, A.M.; Patel, U.K.; O’Connell, K.L.; Dias, D.P.; Moritz, M.; Agard, D.; Stults, J.T.; Stearns, T. Gcp5 and gcp6: Two new members of the human gamma-tubulin complex. Mol. Biol. Cell 2001, 12, 3340–3352. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Masuda, H.; Horii, J.; Kuma, K.; Yokoyama, N.; Ohba, T.; Nishitani, H.; Miyata, T.; Tanaka, M.; Nishimoto, T. When overexpressed, a novel centrosomal protein, ranbpm, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 1998, 143, 1041–1052. [Google Scholar] [CrossRef]
- Negi, S.S.; Olson, M.O. Effects of interphase and mitotic phosphorylation on the mobility and location of nucleolar protein b23. J. Cell Sci. 2006, 119, 3676–3685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niethammer, M.; Smith, D.S.; Ayala, R.; Peng, J.; Ko, J.; Lee, M.S.; Morabito, M.; Tsai, L.H. Nudel is a novel cdk5 substrate that associates with lis1 and cytoplasmic dynein. Neuron 2000, 28, 697–711. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, T.; Takahashi, M.; Kim, H.S.; Mukai, H.; Ono, Y. Centrosome-targeting region of cg-nap causes centrosome amplification by recruiting cyclin e-cdk2 complex. Genes Cells 2005, 10, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Horn, H.F.; Tarapore, P.; Tokuyama, Y.; Smulian, A.G.; Chan, P.K.; Knudsen, E.S.; Hofmann, I.A.; Snyder, J.D.; Bove, K.E.; et al. Nucleophosmin/b23 is a target of cdk2/cyclin e in centrosome duplication. Cell 2000, 103, 127–140. [Google Scholar] [CrossRef] [Green Version]
- Ou, Y.Y.; Mack, G.J.; Zhang, M.; Rattner, J.B. Cep110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 2002, 115, 1825–1835. [Google Scholar]
- Ouchi, M.; Fujiuchi, N.; Sasai, K.; Katayama, H.; Minamishima, Y.A.; Ongusaha, P.P.; Deng, C.; Sen, S.; Lee, S.W.; Ouchi, T. Brca1 phosphorylation by aurora-a in the regulation of g2 to m transition. J. Biol. Chem. 2004, 279, 19643–19648. [Google Scholar] [CrossRef] [PubMed]
- Piehl, M.; Cassimeris, L. Organization and dynamics of growing microtubule plus ends during early mitosis. Mol. Biol. Cell 2003, 14, 916–925. [Google Scholar] [CrossRef] [Green Version]
- Pietromonaco, S.F.; Seluja, G.A.; Elias, L. Identification of enzymatically active ca2+/calmodulin-dependent protein kinase in centrosomes of hemopoietic cells. Blood Cells Mol. Dis. 1995, 21, 34–41. [Google Scholar] [CrossRef]
- Pockwinse, S.M.; Krockmalnic, G.; Doxsey, S.J.; Nickerson, J.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Penman, S. Cell cycle independent interaction of cdc2 with the centrosome, which is associated with the nuclear matrix-intermediate filament scaffold. Proc. Natl. Acad. Sci. USA 1997, 94, 3022–3027. [Google Scholar] [CrossRef] [Green Version]
- Pugacheva, E.N.; Jablonski, S.A.; Hartman, T.R.; Henske, E.P.; Golemis, E.A. Hef1-dependent aurora a activation induces disassembly of the primary cilium. Cell 2007, 129, 1351–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purohit, A.; Tynan, S.H.; Vallee, R.; Doxsey, S.J. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 1999, 147, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Rapley, J.; Baxter, J.E.; Blot, J.; Wattam, S.L.; Casenghi, M.; Meraldi, P.; Nigg, E.A.; Fry, A.M. Coordinate regulation of the mother centriole component nlp by nek2 and plk1 protein kinases. Mol. Cell. Biol. 2005, 25, 1309–1324. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues-Martins, A.; Riparbelli, M.; Callaini, G.; Glover, D.M.; Bettencourt-Dias, M. Revisiting the role of the mother centriole in centriole biogenesis. Science 2007, 316, 1046–1050. [Google Scholar] [CrossRef] [Green Version]
- Rosales, J.L.; Rattner, J.B.; Lee, K.Y. Cdk5 in the centriolar appendages mediates cenexin1 localization and primary cilia formation. Cell Cycle 2010, 9, 2037–2039. [Google Scholar] [CrossRef] [Green Version]
- Ruffner, H.; Jiang, W.; Craig, A.G.; Hunter, T.; Verma, I.M. Brca1 is phosphorylated at serine 1497 in vivo at a cyclin-dependent kinase 2 phosphorylation site. Mol. Cell. Biol. 1999, 19, 4843–4854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, F.; Krzywicka, A.; Klotz, C.; Keller, A.; Cohen, J.; Koll, F.; Balavoine, G.; Beisson, J. The sm19 gene, required for duplication of basal bodies in paramecium, encodes a novel tubulin, eta-tubulin. Curr. Biol. 2000, 10, 1451–1454. [Google Scholar] [CrossRef] [Green Version]
- Saavedra, H.I.; Maiti, B.; Timmers, C.; Altura, R.; Tokuyama, Y.; Fukasawa, K.; Leone, G. Inactivation of e2f3 results in centrosome amplification. Cancer Cell 2003, 3, 333–346. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, S.; Shionoya, A.; Ishida, M.; Gambello, M.J.; Yingling, J.; Wynshaw-Boris, A.; Hirotsune, S. A lis1/nudel/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 2000, 28, 681–696. [Google Scholar] [CrossRef] [Green Version]
- Schafer, D.A.; Gill, S.R.; Cooper, J.A.; Heuser, J.E.; Schroer, T.A. Ultrastructural analysis of the dynactin complex: An actin-related protein is a component of a filament that resembles f-actin. J. Cell Biol. 1994, 126, 403–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroder, J.M.; Schneider, L.; Christensen, S.T.; Pedersen, L.B. Eb1 is required for primary cilia assembly in fibroblasts. Curr. Biol. 2007, 17, 1134–1139. [Google Scholar] [CrossRef] [Green Version]
- Sedjai, F.; Acquaviva, C.; Chevrier, V.; Chauvin, J.P.; Coppin, E.; Aouane, A.; Coulier, F.; Tolun, A.; Pierres, M.; Birnbaum, D.; et al. Control of ciliogenesis by for20, a novel centrosome and pericentriolar satellite protein. J. Cell Sci. 2010, 123, 2391–2401. [Google Scholar] [CrossRef] [Green Version]
- Shinmura, K.; Bennett, R.A.; Tarapore, P.; Fukasawa, K. Direct evidence for the role of centrosomally localized p53 in the regulation of centrosome duplication. Oncogene 2007, 26, 2939–2944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siller, S.S.; Sharma, H.; Li, S.; Yang, J.; Zhang, Y.; Holtzman, M.J.; Winuthayanon, W.; Colognato, H.; Holdener, B.C.; Li, F.Q.; et al. Conditional knockout mice for the distal appendage protein cep164 reveal its essential roles in airway multiciliated cell differentiation. PLoS Genet. 2017, 13, e1007128. [Google Scholar] [CrossRef]
- Sillibourne, J.E.; Milne, D.M.; Takahashi, M.; Ono, Y.; Meek, D.W. Centrosomal anchoring of the protein kinase ck1delta mediated by attachment to the large, coiled-coil scaffolding protein cg-nap/akap450. J. Mol. Biol. 2002, 322, 785–797. [Google Scholar] [CrossRef]
- Sir, J.H.; Barr, A.R.; Nicholas, A.K.; Carvalho, O.P.; Khurshid, M.; Sossick, A.; Reichelt, S.; D’Santos, C.; Woods, C.G.; Gergely, F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 2011, 43, 1147–1153. [Google Scholar] [CrossRef] [Green Version]
- Sonn, S.; Oh, G.T.; Rhee, K. Nek2 and its substrate, centrobin/nip2, are required for proper meiotic spindle formation of the mouse oocytes. Zygote 2011, 19, 15–20. [Google Scholar] [CrossRef]
- Soung, N.K.; Park, J.E.; Yu, L.R.; Lee, K.H.; Lee, J.M.; Bang, J.K.; Veenstra, T.D.; Rhee, K.; Lee, K.S. Plk1-dependent and -independent roles of an odf2 splice variant, hcenexin1, at the centrosome of somatic cells. Dev. Cell 2009, 16, 539–550. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y. P53 and its downstream proteins as molecular targets of cancer. Mol. Carcinog. 2006, 45, 409–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sydor, A.M.; Coyaud, E.; Rovelli, C.; Laurent, E.; Liu, H.; Raught, B.; Mennella, V. PPP1R35 is a novel centrosomal protein that regulates centriole length in concert with the microcephaly protein rttn. eLife 2018, 7, e37846. [Google Scholar] [CrossRef] [PubMed]
- Szebenyi, G.; Hall, B.; Yu, R.; Hashim, A.I.; Kramer, H. Hook2 localizes to the centrosome, binds directly to centriolin/cep110 and contributes to centrosomal function. Traffic 2007, 8, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Mukai, H.; Oishi, K.; Isagawa, T.; Ono, Y. Association of immature hypophosphorylated protein kinase cepsilon with an anchoring protein cg-nap. J. Biol. Chem. 2000, 275, 34592–34596. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.; Shibata, H.; Shimakawa, M.; Miyamoto, M.; Mukai, H.; Ono, Y. Characterization of a novel giant scaffolding protein, cg-nap, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J. Biol. Chem. 1999, 274, 17267–17274. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.; Yamagiwa, A.; Nishimura, T.; Mukai, H.; Ono, Y. Centrosomal proteins cg-nap and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell 2002, 13, 3235–3245. [Google Scholar] [CrossRef] [Green Version]
- Tanos, B.E.; Yang, H.J.; Soni, R.; Wang, W.J.; Macaluso, F.P.; Asara, J.M.; Tsou, M.F. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013, 27, 163–168. [Google Scholar] [CrossRef] [Green Version]
- Terada, Y.; Uetake, Y.; Kuriyama, R. Interaction of aurora-a and centrosomin at the microtubule-nucleating site in drosophila and mammalian cells. J. Cell Biol. 2003, 162, 757–763. [Google Scholar] [CrossRef]
- Toya, M.; Terasawa, M.; Nagata, K.; Iida, Y.; Sugimoto, A. A kinase-independent role for aurora a in the assembly of mitotic spindle microtubules in caenorhabditis elegans embryos. Nat. Cell Biol. 2011, 13, 708–714. [Google Scholar] [CrossRef]
- Tsai, J.J.; Hsu, W.B.; Liu, J.H.; Chang, C.W.; Tang, T.K. Author correction: Cep120 interacts with c2cd3 and talpid3 and is required for centriole appendage assembly and ciliogenesis. Sci. Rep. 2020, 10, 1265. [Google Scholar] [CrossRef]
- Tsang, W.Y.; Spektor, A.; Luciano, D.J.; Indjeian, V.B.; Chen, Z.; Salisbury, J.L.; Sanchez, I.; Dynlacht, B.D. Cp110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol. Biol. Cell 2006, 17, 3423–3434. [Google Scholar] [CrossRef] [Green Version]
- Tzivion, G.; Shen, Y.H.; Zhu, J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 2001, 20, 6331–6338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzbekov, R.; Prigent, C.; Arlot-Bonnemains, Y. Cell cycle analysis and synchronization of the xenopus laevis xl2 cell line: Study of the kinesin related protein xleg5. Microsc. Res. Tech. 1999, 45, 31–42. [Google Scholar] [CrossRef]
- Uzbekov, R.E. Dynamics of Functional Activity of the Centrosome during Cell Cycle. Ph.D. Thesis, Moscow State University, Moscow, Russia, 2004. [Google Scholar]
- Vasquez, R.J.; Gard, D.L.; Cassimeris, L. Phosphorylation by cdk1 regulates xmap215 function in vitro. Cell Motil. Cytoskelet. 1999, 43, 310–321. [Google Scholar] [CrossRef]
- Vaughan, K.T.; Vallee, R.B. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150glued. J. Cell Biol. 1995, 131, 1507–1516. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Shao, N.; Ding, Q.M.; Cui, J.; Reddy, E.S.; Rao, V.N. Brca1 proteins are transported to the nucleus in the absence of serum and splice variants brca1a, brca1b are tyrosine phosphoproteins that associate with e2f, cyclins and cyclin dependent kinases. Oncogene 1997, 15, 143–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.F.; Takenaka, K.; Nakanishi, A.; Miki, Y. Brca2 and nucleophosmin coregulate centrosome amplification and form a complex with the rho effector kinase rock2. Cancer Res. 2011, 71, 68–77. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Failler, M.; Fu, W.; Dynlacht, B.D. A distal centriolar protein network controls organelle maturation and asymmetry. Nat. Commun. 2018, 9, 3938. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.H.; Mayer, B.; Stemmann, O.; Nigg, E.A. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci. 2010, 123, 806–813. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Takao, D.; Ito, K.K.; Takahashi, M.; Kitagawa, D. The cep57-pericentrin module organizes pcm expansion and centriole engagement. Nat. Commun. 2019, 10, 931. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Xu, Q.; Zhang, Y.; Li, Y.; Zhang, Q.; Hu, Z.; Harris, P.C.; Torres, V.E.; Ling, K.; Hu, J. Transition fibre protein fbf1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat. Commun. 2013, 4, 2750. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Kim, T.S.; Ahn, J.I.; Meng, L.; Chen, Y.; Ryu, E.K.; Ku, B.; Zhou, M.; Kim, S.J.; Bang, J.K.; et al. Requirement of the cep57-cep63 interaction for proper cep152 recruitment and centriole duplication. Mol. Cell. Biol. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Timmers, C.; Maiti, B.; Saavedra, H.I.; Sang, L.; Chong, G.T.; Nuckolls, F.; Giangrande, P.; Wright, F.A.; Field, S.J.; et al. The e2f1-3 transcription factors are essential for cellular proliferation. Nature 2001, 414, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Fu, C.; Ding, X.; Guo, Z.; Zenreski, A.; Chen, Y.; Ahmed, K.; Liao, J.; Dou, Z.; Yao, X. Nek2a kinase regulates the localization of numatrin to centrosome in mitosis. FEBS Lett. 2004, 575, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Yarden, R.I.; Brody, L.C. Brca1 interacts with components of the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 1999, 96, 4983–4988. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Zeng, H.; Ning, G.; Reiter, J.F.; Liu, A. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA 2014, 111, 2164–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zach, F.; Grassmann, F.; Langmann, T.; Sorusch, N.; Wolfrum, U.; Stohr, H. The retinitis pigmentosa 28 protein fam161a is a novel ciliary protein involved in intermolecular protein interaction and microtubule association. Hum. Mol. Genet. 2012, 21, 4573–4586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Somasundaram, K.; Peng, Y.; Tian, H.; Zhang, H.; Bi, D.; Weber, B.L.; El-Deiry, W.S. Brca1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 1998, 16, 1713–1721. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Megraw, T.L. Proper recruitment of gamma-tubulin and d-tacc/msps to embryonic drosophila centrosomes requires centrosomin motif 1. Mol. Biol. Cell 2007, 18, 4037–4049. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Fletcher, L.; Muschel, R.J. The role of polo-like kinase 1 in the inhibition of centrosome separation after ionizing radiation. J. Biol. Chem. 2005, 280, 42994–42999. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Jin, S.; Song, Y.; Zhan, Q. Cdc2/cyclin b1 regulates centrosomal nlp proteolysis and subcellular localization. Cancer Biol. Ther. 2010, 10, 945–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmerman, W.C.; Sillibourne, J.; Rosa, J.; Doxsey, S.J. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 2004, 15, 3642–3657. [Google Scholar] [CrossRef] [PubMed]
- Avidor-Reiss, T.; Mazur, M.; Fishman, E.L.; Sindhwani, P. The role of sperm centrioles in human reproduction—The known and the unknown. Front. Cell Dev. Biol. 2019, 7, 188. [Google Scholar] [CrossRef]
- Avidor-Reiss, T.; Turner, K. The evolution of centriole structure: Heterochrony, neoteny, and hypermorphosis. Results Probl. Cell Differ. 2019, 67, 3–15. [Google Scholar] [PubMed]
- Uzbekov, R.E.; Alieva, I.B. The centrosome—A riddle of the “cell processor”. Tsitologiia 2008, 50, 91–112. [Google Scholar] [PubMed]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uzbekov, R.E.; Avidor-Reiss, T. Principal Postulates of Centrosomal Biology. Version 2020. Cells 2020, 9, 2156. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9102156
Uzbekov RE, Avidor-Reiss T. Principal Postulates of Centrosomal Biology. Version 2020. Cells. 2020; 9(10):2156. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9102156
Chicago/Turabian StyleUzbekov, Rustem E., and Tomer Avidor-Reiss. 2020. "Principal Postulates of Centrosomal Biology. Version 2020" Cells 9, no. 10: 2156. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9102156