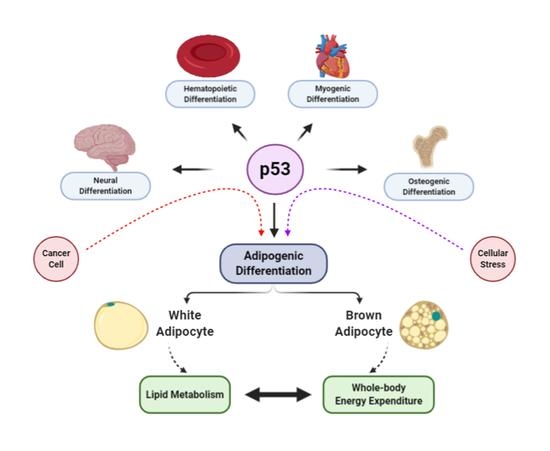
The Intricate Role of p53 in Adipocyte Differentiation and Function
Abstract
:1. Introduction
2. Role of P53 and White Adipocyte Differentiation and Lipid Metabolism
3. Role of P53 in Brown Adipocyte Differentiation and Thermogenesis
4. Role of P53 in Beige Adipocytes and Browning
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Finlay, C.A.; Hinds, P.W.; Levine, A.J. The p53 proto-oncogene can act as a suppressor of transformation. Cell 1989, 57, 1083–1093. [Google Scholar] [CrossRef]
- Horn, H.F.; Vousden, K.H. Coping with stress: Multiple ways to activate p53. Oncogene 2007, 26, 1306–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashcroft, M.; Kubbutat, M.H.; Vousden, K.H. Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 1999, 19, 1751–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olovnikov, I.A.; Kravchenko, J.E.; Chumakov, P.M. Homeostatic functions of the p53 tumor suppressor: Regulation of energy metabolism and antioxidant defense. Semin. Cancer Biol. 2009, 19, 32–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, R.G.; Plas, D.R.; Kubek, S.; Buzzai, M.; Mu, J.; Xu, Y.; Birnbaum, M.J.; Thompson, C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 2005, 18, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.M.; Razmara, M.; Nguyen, D.; Donahue, R.J.; Wubah, J.A.; Knudsen, T.B. Altered expression of mitochondrial 16S ribosomal RNA in p53-deficient mouse embryos revealed by differential display. Biochim. Biophys. Acta 1998, 1403, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Sung, H.J.; Park, J.Y.; Matoba, S.; Hwang, P.M. A pivotal role for p53: Balancing aerobic respiration and glycolysis. J. Bioenerg. Biomembr. 2007, 39, 243–246. [Google Scholar] [CrossRef]
- Buzzai, M.; Jones, R.G.; Amaravadi, R.K.; Lum, J.J.; DeBerardinis, R.J.; Zhao, F.; Viollet, B.; Thompson, C.B. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007, 67, 6745–6752. [Google Scholar] [CrossRef] [Green Version]
- Buckbinder, L.; Talbott, R.; Velasco-Miguel, S.; Takenaka, I.; Faha, B.; Seizinger, B.R.; Kley, N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature 1995, 377, 646–649. [Google Scholar] [CrossRef]
- Polyak, K.; Xia, Y.; Zweier, J.L.; Kinzler, K.W.; Vogelstein, B. A model for p53-induced apoptosis. Nature 1997, 389, 300–305. [Google Scholar] [CrossRef]
- Yoon, K.A.; Nakamura, Y.; Arakawa, H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J. Hum. Genet. 2004, 49, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Feng, Z.; Teresky, A.K.; Levine, A.J. p53 regulates maternal reproduction through LIF. Nature 2007, 450, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.F.; Kaufman, M.H.; Harrison, D.J.; Clarke, A.R. High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 1995, 5, 931–936. [Google Scholar] [CrossRef] [Green Version]
- Matheu, A.; Maraver, A.; Serrano, M. The Arf/p53 pathway in cancer and aging. Cancer Res. 2008, 68, 6031–6034. [Google Scholar] [CrossRef] [Green Version]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef]
- Lavin, M.F.; Gueven, N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006, 13, 941–950. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Soga, T. Cancer metabolism: Key players in metabolic reprogramming. Cancer Sci. 2013, 104, 275–281. [Google Scholar] [CrossRef]
- Schmid, P.; Lorenz, A.; Hameister, H.; Montenarh, M. Expression of p53 during mouse embryogenesis. Development 1991, 113, 857–865. [Google Scholar]
- Tedeschi, A.; Di Giovanni, S. The non-apoptotic role of p53 in neuronal biology: Enlightening the dark side of the moon. EMBO Rep. 2009, 10, 576–583. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Cohen, P.; Spiegelman, B.M. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev. 2013, 27, 234–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Kang, H.J.; Kang, S.I.; Lee, J.E.; Hur, J.; Ge, K.; Mueller, E.; Li, H.; Lee, B.C.; Lee, S.B. A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev. Cell 2013, 26, 393–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Hur, W.; Lee, S.B. Intricate Transcriptional Networks of Classical Brown and Beige Fat Cells. Front. Endocrinol. (Lausanne) 2015, 6, 124. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 2016, 17, 691–702. [Google Scholar] [CrossRef]
- Han, J.; Lee, J.E.; Jin, J.; Lim, J.S.; Oh, N.; Kim, K.; Chang, S.I.; Shibuya, M.; Kim, H.; Koh, G.Y. The spatiotemporal development of adipose tissue. Development 2011, 138, 5027–5037. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef]
- Chau, Y.Y.; Bandiera, R.; Serrels, A.; Martinez-Estrada, O.M.; Qing, W.; Lee, M.; Slight, J.; Thornburn, A.; Berry, R.; McHaffie, S.; et al. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 2014, 16, 367–375. [Google Scholar] [CrossRef] [Green Version]
- Di Girolamo, M.; Fine, J.B.; Tagra, K.; Rossmanith, R. Qualitative regional differences in adipose tissue growth and cellularity in male Wistar rats fed ad libitum. Am. J. Physiol. 1998, 274, R1460–R1467. [Google Scholar] [CrossRef]
- Liu, S.; Kim, T.H.; Franklin, D.A.; Zhang, Y. Protection against High-Fat-Diet-Induced Obesity in MDM2(C305F) Mice Due to Reduced p53 Activity and Enhanced Energy Expenditure. Cell Rep. 2017, 18, 1005–1018. [Google Scholar] [CrossRef]
- Yahagi, N.; Shimano, H.; Matsuzaka, T.; Najima, Y.; Sekiya, M.; Nakagawa, Y.; Ide, T.; Tomita, S.; Okazaki, H.; Tamura, Y.; et al. p53 Activation in adipocytes of obese mice. J. Biol. Chem. 2003, 278, 25395–25400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molchadsky, A.; Ezra, O.; Amendola, P.G.; Krantz, D.; Kogan-Sakin, I.; Buganim, Y.; Rivlin, N.; Goldfinger, N.; Folgiero, V.; Falcioni, R.; et al. p53 is required for brown adipogenic differentiation and has a protective role against diet-induced obesity. Cell Death Differ. 2013, 20, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Molchadsky, A.; Shats, I.; Goldfinger, N.; Pevsner-Fischer, M.; Olson, M.; Rinon, A.; Tzahor, E.; Lozano, G.; Zipori, D.; Sarig, R.; et al. p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS ONE 2008, 3, e3707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Liu, M.; Du, X.; Zhang, R.; Xue, Y.; Zhang, Y.; Zhu, W.; Li, D.; Zhao, A.; Liu, Y. Role of p53 in preadipocyte differentiation. Cell Biol. Int. 2014, 38, 1384–1393. [Google Scholar] [CrossRef]
- Chang, E.; Kim, C.Y. Natural Products and Obesity: A Focus on the Regulation of Mitotic Clonal Expansion during Adipogenesis. Molecules 2019, 24, 1157. [Google Scholar] [CrossRef] [Green Version]
- Naaz, A.; Holsberger, D.R.; Iwamoto, G.A.; Nelson, A.; Kiyokawa, H.; Cooke, P.S. Loss of cyclin-dependent kinase inhibitors produces adipocyte hyperplasia and obesity. FASEB J. 2004, 18, 1925–1927. [Google Scholar] [CrossRef]
- Inoue, N.; Yahagi, N.; Yamamoto, T.; Ishikawa, M.; Watanabe, K.; Matsuzaka, T.; Nakagawa, Y.; Takeuchi, Y.; Kobayashi, K.; Takahashi, A.; et al. Cyclin-dependent kinase inhibitor, p21WAF1/CIP1, is involved in adipocyte differentiation and hypertrophy, linking to obesity, and insulin resistance. J. Biol. Chem. 2008, 283, 21220–21229. [Google Scholar] [CrossRef] [Green Version]
- Hallenborg, P.; Petersen, R.K.; Feddersen, S.; Sundekilde, U.; Hansen, J.B.; Blagoev, B.; Madsen, L.; Kristiansen, K. PPARgamma ligand production is tightly linked to clonal expansion during initiation of adipocyte differentiation. J. Lipid Res. 2014, 55, 2491–2500. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.Q.; Gronborg, M.; Huang, H.; Kim, J.W.; Otto, T.C.; Pandey, A.; Lane, M.D. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 9766–9771. [Google Scholar] [CrossRef] [Green Version]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Evan, D.; Rosen, C.J.W.; Puigserver, P.; Spiegelman, B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000, 14, 1293–1307. [Google Scholar]
- Rosen, E.D.; Spiegelman, B.M. What We Talk About When We Talk About Fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okita, N.; Ishikawa, N.; Mizunoe, Y.; Oku, M.; Nagai, W.; Suzuki, Y.; Matsushima, S.; Mikami, K.; Okado, H.; Sasaki, T.; et al. Inhibitory effect of p53 on mitochondrial content and function during adipogenesis. Biochem. Biophys. Res. Commun. 2014, 446, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yang, X.; Zhao, Q.; Li, Z.; Fu, F.; Zhang, H.; Zheng, M.; Zhang, S. Molecular Mechanism of Stem Cell Differentiation into Adipocytes and Adipocyte Differentiation of Malignant Tumor. Stem Cells Int. 2020, 2020, 8892300. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Serpell, C.J.; Fong, I.L.; Wong, E.H. Molecular Mechanisms of Adipogenesis: The Anti-adipogenic Role of AMP-Activated Protein Kinase. Front. Mol. Biosci. 2020, 7, 76. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [Green Version]
- Hallenborg, P.; Fjaere, E.; Liaset, B.; Petersen, R.K.; Murano, I.; Sonne, S.B.; Falkerslev, M.; Winther, S.; Jensen, B.A.; Ma, T.; et al. p53 regulates expression of uncoupling protein 1 through binding and repression of PPARgamma coactivator-1alpha. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E116–E128. [Google Scholar] [CrossRef] [Green Version]
- Yadav, N.; Cheng, D.; Richard, S.; Morel, M.; Iyer, V.R.; Aldaz, C.M.; Bedford, M.T. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008, 9, 193–198. [Google Scholar] [CrossRef]
- Behera, A.K.; Bhattacharya, A.; Vasudevan, M.; Kundu, T.K. p53 mediated regulation of coactivator associated arginine methyltransferase 1 (CARM1) expression is critical for suppression of adipogenesis. FEBS J. 2018, 285, 1730–1744. [Google Scholar] [CrossRef] [Green Version]
- Boregowda, S.V.; Krishnappa, V.; Strivelli, J.; Haga, C.L.; Booker, C.N.; Phinney, D.G. Basal p53 expression is indispensable for mesenchymal stem cell integrity. Cell Death Differ. 2018, 25, 679–692. [Google Scholar] [CrossRef] [Green Version]
- Berberich, S.J.; Litteral, V.; Mayo, L.D.; Tabesh, D.; Morris, D. mdm-2 gene amplification in 3T3-L1 preadipocytes. Differentiation 1999, 64, 205–212. [Google Scholar] [CrossRef]
- Liu, Z.; Jin, L.; Yang, J.K.; Wang, B.; Wu, K.K.L.; Hallenborg, P.; Xu, A.; Cheng, K.K.Y. The Dysfunctional MDM2-p53 Axis in Adipocytes Contributes to Aging-Related Metabolic Complications by Induction of Lipodystrophy. Diabetes 2018, 67, 2397–2409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallenborg, P.; Siersbaek, M.; Barrio-Hernandez, I.; Nielsen, R.; Kristiansen, K.; Mandrup, S.; Grontved, L.; Blagoev, B. MDM2 facilitates adipocyte differentiation through CRTC-mediated activation of STAT3. Cell Death Dis. 2016, 7, e2289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallenborg, P.; Feddersen, S.; Francoz, S.; Murano, I.; Sundekilde, U.; Petersen, R.K.; Akimov, V.; Olson, M.V.; Lozano, G.; Cinti, S.; et al. Mdm2 controls CREB-dependent transactivation and initiation of adipocyte differentiation. Cell Death Differ. 2012, 19, 1381–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimmel, A.R.; Sztalryd, C. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu. Rev. Nutr. 2016, 36, 471–509. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Sohn, J.H.; Han, J.S.; Park, Y.J.; Jeon, Y.G.; Ji, Y.; Dalen, K.T.; Sztalryd, C.; Kimmel, A.R.; Kim, J.B. Perilipin 3 Deficiency Stimulates Thermogenic Beige Adipocytes Through PPARalpha Activation. Diabetes 2018, 67, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Nerstedt, A.; Smith, U. Reduced subcutaneous adipogenesis in human hypertrophic obesity is linked to senescent precursor cells. Nat. Commun. 2019, 10, 2757. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, I.; Yoshida, Y.; Katsuno, T.; Tateno, K.; Okada, S.; Moriya, J.; Yokoyama, M.; Nojima, A.; Ito, T.; Zechner, R.; et al. p53-induced adipose tissue inflammation is critically involved in the development of insulin resistance in heart failure. Cell Metab. 2012, 15, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Schupp, M.; Chen, F.; Briggs, E.R.; Rao, S.; Pelzmann, H.J.; Pessentheiner, A.R.; Bogner-Strauss, J.G.; Lazar, M.A.; Baldwin, D.; Prokesch, A. Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues. BMC Genom. 2013, 14, 758. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xu, M.; Peng, Y.; Naren, Q.; Xu, Y.; Wang, X.; Yang, G.; Shi, X.; Li, X. Triptolide enhances lipolysis of adipocytes by enhancing ATGL transcription via upregulation of p53. Phytother. Res. 2020. [Google Scholar] [CrossRef]
- Kung, C.P.; Leu, J.I.; Basu, S.; Khaku, S.; Anokye-Danso, F.; Liu, Q.; George, D.L.; Ahima, R.S.; Murphy, M.E. The P72R Polymorphism of p53 Predisposes to Obesity and Metabolic Dysfunction. Cell Rep. 2016, 14, 2413–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derdak, Z.; Villegas, K.A.; Harb, R.; Wu, A.M.; Sousa, A.; Wands, J.R. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 785–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handschin, C.; Spiegelman, B.M. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006, 27, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S. The adipose organ. Prostaglandins Leukot. Essent. Fat. Acids 2005, 73, 9–15. [Google Scholar] [CrossRef]
- Hirning, U.; Schmid, P.; Schulz, W.A.; Kozak, L.P.; Hameister, H. In developing brown adipose tissue c-myc protooncogene expression is restricted to early differentiation stages. Cell Differ. Dev. 1989, 27, 243–248. [Google Scholar] [CrossRef]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef] [Green Version]
- Harms, M.J.; Ishibashi, J.; Wang, W.; Lim, H.W.; Goyama, S.; Sato, T.; Kurokawa, M.; Won, K.J.; Seale, P. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 2014, 19, 593–604. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, J.; Seale, P. Functions of Prdm16 in thermogenic fat cells. Temperature (Austin) 2015, 2, 65–72. [Google Scholar] [CrossRef]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [Green Version]
- Westerberg, R.; Mansson, J.E.; Golozoubova, V.; Shabalina, I.G.; Backlund, E.C.; Tvrdik, P.; Retterstol, K.; Capecchi, M.R.; Jacobsson, A. ELOVL3 is an important component for early onset of lipid recruitment in brown adipose tissue. J. Biol. Chem. 2006, 281, 4958–4968. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Yon Toh, S.; Chen, Z.; Guo, K.; Ng, C.P.; Ponniah, S.; Lin, S.C.; Hong, W.; Li, P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat. Genet. 2003, 35, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Barbera, M.J.; Schluter, A.; Pedraza, N.; Iglesias, R.; Villarroya, F.; Giralt, M. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 2001, 276, 1486–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jash, S.; Banerjee, S.; Lee, M.J.; Farmer, S.R.; Puri, V. CIDEA Transcriptionally Regulates UCP1 for Britening and Thermogenesis in Human Fat Cells. iScience 2019, 20, 73–89. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Fernand, V.E.; Henagan, T.M.; Shin, J.; Huypens, P.; Newman, S.; Gettys, T.W.; Chang, J.S. Regulation of Brown and White Adipocyte Transcriptome by the Transcriptional Coactivator NT-PGC-1alpha. PLoS ONE 2016, 11, e0159990. [Google Scholar] [CrossRef]
- Gan, L.; Liu, Z.; Jin, W.; Zhou, Z.; Sun, C. Foxc2 enhances proliferation and inhibits apoptosis through activating Akt/mTORC1 signaling pathway in mouse preadipocytes. J. Lipid Res. 2015, 56, 1471–1480. [Google Scholar] [CrossRef] [Green Version]
- Hader, C.; Marlier, A.; Cantley, L. Mesenchymal-epithelial transition in epithelial response to injury: The role of Foxc2. Oncogene 2010, 29, 1031–1040. [Google Scholar] [CrossRef] [Green Version]
- Cederberg, A.; Gronning, L.M.; Ahren, B.; Tasken, K.; Carlsson, P.; Enerback, S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell 2001, 106, 563–573. [Google Scholar] [CrossRef] [Green Version]
- Al-Massadi, O.; Porteiro, B.; Kuhlow, D.; Köhler, M.; Gonzalez-Rellan, M.J.; Garcia-Lavandeira, M.; Díaz-Rodríguez, E.; Quiñones, M.; Senra, A.; Alvarez, C.V.; et al. Pharmacological and Genetic Manipulation of p53 in Brown Fat at Adult But Not Embryonic Stages Regulates Thermogenesis and Body Weight in Male Mice. Endocrinology 2016, 157, 2735–2749. [Google Scholar] [CrossRef] [Green Version]
- Minamino, T.; Orimo, M.; Shimizu, I.; Kunieda, T.; Yokoyama, M.; Ito, T.; Nojima, A.; Nabetani, A.; Oike, Y.; Matsubara, H.; et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat. Med. 2009, 15, 1082–1087. [Google Scholar] [CrossRef]
- Fu, W.; Liu, Y.; Sun, C.; Yin, H. Transient p53 inhibition sensitizes aged white adipose tissue for beige adipocyte recruitment by blocking mitophagy. FASEB J. 2019, 33, 844–856. [Google Scholar] [CrossRef]
- Lozano, G. Mouse models of p53 functions. Cold Spring Harb. Perspect. Biol. 2010, 2, a001115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Stiewe, T. The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer 2007, 7, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Pfitzenmaier, J.; Vessella, R.; Higano, C.S.; Noteboom, J.L.; Wallace, D., Jr.; Corey, E. Elevation of cytokine levels in cachectic patients with prostate carcinoma. Cancer 2003, 97, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petruzzelli, M.; Wagner, E.F. Mechanisms of metabolic dysfunction in cancer-associated cachexia. Genes Dev. 2016, 30, 489–501. [Google Scholar] [CrossRef] [Green Version]
- Gudkov, A.V.; Gurova, K.V.; Komarova, E.A. Inflammation and p53: A Tale of Two Stresses. Genes Cancer 2011, 2, 503–516. [Google Scholar] [CrossRef]
- Filichia, E.; Shen, H.; Zhou, X.; Qi, X.; Jin, K.; Greig, N.; Hoffer, B.; Luo, Y. Forebrain neuronal specific ablation of p53 gene provides protection in a cortical ischemic stroke model. Neuroscience 2015, 295, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Jirkof, P.; Bratcher, N.; Medina, L.; Strasburg, D.; Ebert, P.; Gaskill, B.N. The effect of group size, age and handling frequency on inter-male aggression in CD 1 mice. Sci. Rep. 2020, 10, 2253. [Google Scholar] [CrossRef] [Green Version]
- Espinosa, J.M.; Emerson, B.M. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 2001, 8, 57–69. [Google Scholar] [CrossRef]
- Wei, C.L.; Wu, Q.; Vega, V.B.; Chiu, K.P.; Ng, P.; Zhang, T.; Shahab, A.; Yong, H.C.; Fu, Y.; Weng, Z.; et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006, 124, 207–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajimura, S.; Seale, P.; Kubota, K.; Lunsford, E.; Frangioni, J.V.; Gygi, S.P.; Spiegelman, B.M. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 2009, 460, 1154–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Sun, Y. Targeting p53 for Novel Anticancer Therapy. Transl. Oncol. 2010, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.J.; Li, X.; Hunt, C.; Wang, W.; Wang, H.; Zhang, R. Natural products targeting the p53-MDM2 pathway and mutant p53: Recent advances and implications in cancer medicine. Genes Dis. 2018, 5, 204–219. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.K.; Chung, Y.S.; Lee, J.H.; Chun, J.M.; Park, J.H. The Intricate Role of p53 in Adipocyte Differentiation and Function. Cells 2020, 9, 2621. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9122621
Lee YK, Chung YS, Lee JH, Chun JM, Park JH. The Intricate Role of p53 in Adipocyte Differentiation and Function. Cells. 2020; 9(12):2621. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9122621
Chicago/Turabian StyleLee, Yun Kyung, Yu Seong Chung, Ji Hye Lee, Jin Mi Chun, and Jun Hong Park. 2020. "The Intricate Role of p53 in Adipocyte Differentiation and Function" Cells 9, no. 12: 2621. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9122621