This section contains two subsections, one for Data Mining Experiments and one for Databases Experiments. The tests run on a computer with Windows 10, Intel Core i7-8750H 2.20 GHz processor, 16 GB RAM, and 256 GB SSD. SQL and NoSQL databases evaluated versions are SQL Server version 2017, MongoDB version 4.4, and Cassandra version 3.11.10.
5.1. Data Mining Experiments—Classification Tests
The Orange Data Mining platform performed all classification tests in version 3.2.9. These tests used the table SymptomsCovid because of the interesting features in it. This table has 3,021,444 records and 11 features. The features used for these tests were Gender, Age60orOlder, TestReason, Headache, Fever, Cough, SoreThroat, ShortnessOfBreath, and TestResult.
Classification tests were performed to create a suitable and helpful COVID-19 predictive model. The main goal of this model is to predict patients’ symptoms and patients’ test results based on the data features used. All these tests were performed with the cross-validation technique with ten subsets, nine for training and one for testing. Regarding the parameters of the algorithms, the default values provided by Orange Data Mining were used.
First, a precise test classification result and each patient symptom were carried out, and later the features were concatenated. Concatenation is appending one string to the end of another string.
Description of each singular classification test:
I—Target: TestResult; 3 Possibilities: Negative (0), Positive (1), Inconclusive (2);
II—Target: Cough; 2 Possibilities: Positive (1), Negative (0);
III—Target: Fever; 2 Possibilities: Positive (1), Negative (0);
IV—Target: SoreThroat; 2 Possibilities: Positive (1), Negative (0);
V—Target: ShortnessOfBreath; 2 Possibilities: Positive (1), Negative (0);
VI—Target: Headache; 2 Possibilities: Positive (1), Negative (0).
Description of each concatenated classification test:
VII—Target: CoughTestResult; 6 Possibilities: Positive for cough and Negative for test result (10), Positive for both (11), Positive for cough and Inconclusive for test result (12), Negative for both (00), Negative for cough and Positive for test result (01), Negative for cough and Inconclusive for test result (02);
VIII—Target: FeverTestResult; 6 Possibilities: Positive for fever and Negative for test result (10), Positive for both (11), Positive for fever and Inconclusive for test result (12), Negative for both (00), Negative for fever and Positive for test result (01), Negative for fever and Inconclusive for test result (02);
IX—Target: SoreThroatTestResult; 6 Possibilities: Positive for sore throat and Negative for test result (10), Positive for both (11), Positive for sore throat and Inconclusive for test result (12), Negative for both (00), Negative for sore throat and Positive for test result (01), Negative for sore throat and Inconclusive for test result (02);
X—Target: ShortnessOfBreathTestResult; 6 Possibilities: Positive for shortness of breath and Negative for test result (10), Positive for both (11), Positive for shortness of breath and Inconclusive for test result (12), Negative for both (00), Negative for shortness of breath and Positive for test result (01), Negative for shortness of breath and Inconclusive for test result (02);
XI—Target: HeadacheTestResult; 6 Possibilities: Positive for headache and Negative for test result (10), Positive for both (11), Positive for headache and Inconclusive for test result (12), Negative for both (00), Negative for headache and Positive for test result (01), Negative for headache and Inconclusive for test result (02);
XII—Target: CoughShortnessOfBreathTestResult; 12 Possibilities: Negative for all (000), Negative for cough and shortness of breath and Positive for test result (001), Negative for cough and test result and Positive for shortness of breath (010), Negative for cough and Positive for shortness of breath and test result (011), Positive for cough and Negative for shortness of breath and test result (100), Positive for cough and test result and Negative for shortness of breath (101), Positive for cough and shortness of breath and Negative for test result (110), Positive for all (111), Negative for cough and shortness of breath and Inconclusive for test result (002), Positive for cough, Negative for shortness of breath and Inconclusive for test result (102), Negative for cough, Positive for shortness of breath and Inconclusive for test result (012), Positive for cough and shortness of breath and Inconclusive for test result (112);
XIII—Target: FeverHeadacheTestResult; 12 Possibilities: (000), (001), (010), (011), (100), (101), (110), (111), (002), (102), (012), (112);
XIV—Target: SoreThroatShortnessOfBreathTestResult; 12 Possibilities: (000), (001), (010), (011), (100), (101), (110), (111), (002), (102), (012), (112).
Accuracy, Recall, Precision, and F1 Score were the classification evaluation metrics used in the experiments. Accuracy is the proportion of actual results among the number of the total cases examined:
where TP, FN, FP, and TN represent the number of true positives, false negatives, false positives, and true negatives, respectively.
A recall is the ratio of correctly predicted positive observations to the predicted positive observations:
Precision is the ratio of correctly predicted positive observations to the total predicted positive observations:
F1 Score is the weighted average of Precision and Recall:
Table 1,
Table 2 and
Table 3 show the accuracy, precision, and F1 Score results for the tests with singular targets, respectively. The Recall results were similar to the Accuracy results, and for that reason, a table was not created for Recall. The last column of all tables shows the average of the results (arithmetic mean).
In
Table 1, all algorithms had outstanding accuracies. Random Forest was the best algorithm in all tests, obtaining accuracy between 99.6% when predicting breath shortness and 92.8% when predicting the COVID-19 test result. In some tests, Decision Trees has obtained the same accuracy as Random Forest (III, IV, and V) and Logistic Regression in V. The small decrease in accuracy when the COVID-19 test result is predicted since there are tests with inconclusive results. If the tests were only positive or negative, the accuracy would be higher.
In
Table 2, all algorithms had remarkable precisions. Naïve Bayes was the best algorithm in four of six (III, IV, V, VI), and Random Forest was the best in 2 of 6 tests (I and II). The test with the best precision predicted breath shortness with 99.4%.
In
Table 3, all algorithms had a great F1 Score and other metrics. Random Forest was the best algorithm in three of six tests (I, II, and V) and Logistic Regression (IV, V, VI). Naïve Bayes was the best in the test III. The test with the best F1 Score was when predicting breath shortness with 99.4%.
This first singular test set calculated average accuracy, precision, and F1 Score for each algorithm to select the best. The best algorithm was Random Forest, with an average accuracy of 96.88%, an average precision of 95.97%, and an average F1 Score of 96.15%. Second, Logistic Regression with an average accuracy of 96.77%, an average precision of 95.6%, and an average F1 Score of 95.87%. In third, the Decision Tree with an average accuracy of 96.60%, an average precision of 95.17%, and an average F1 Score of 95.85%. Finally, in fourth, Naïve Bayes with an average accuracy of 95.28%, an average precision of 95.73%, and an average F1 Score of 95.43%.
Table 4,
Table 5 and
Table 6 show the accuracy, precision, and F1 Score results for the tests with concatenated targets, respectively. It should be stated that
Table 1 and
Table 4 show the accuracy results.
Table 2 and
Table 5 show the precision results.
Table 3 and
Table 6 show the F1 Score results for the singular and concatenated classification tests made.
In
Table 4, all algorithms show significant accuracies. The small decrease in this test is because fewer features predict more concatenated features. Despite that, the accuracy remained high. In all tests, Random Forest was the best algorithm, obtaining an accuracy between 92.4% when predicting shortness of breath combined with the test result and 89.0% when predicting the fever combined with headache and test result. Decision Trees has obtained the same accuracy as Random Forest (VII, VIII, X, XI, XII, XIII, and XIV).
In
Table 5, all algorithms show good precisions. Random Forest and Decision Tree were the best algorithms in 6 of 8 tests (VII, IX, X, XI, XII, and XIV), and Naïve Bayes was the best in two of eight tests (VIII and XIII). The test with the best precision was when predicting breath shortness concatenated with a test result of 90.4%.
In
Table 6, all algorithms had a good F1 Score. Random Forest and Decision Tree were the best algorithms in 6 of 8 tests (VII, IX, X, XI, XII, and XIV), and Naïve Bayes was the best in two of eight tests (VII and XII). The test with the best F1 Score was when predicting breath shortness, concatenated with a test result of 91.3%.
This second concatenated test set calculated average accuracy, precision, and F1 Score for each algorithm to select the best. The best algorithm was Random Forest, with an average accuracy of 90.68%, an average precision of 87.95%, and an average F1 Score of 89.03%. In second, Decision Tree with an average accuracy of 90.66%, an average precision of 87.95%, and an average F1 Score of 89.02%. In third, Naïve Bayes with an average accuracy of 89.91%, an average precision of 87.16%, and an average F1 Score of 88.39%. Finally, in fourth, Logistic Regression, with an average accuracy of 90.15%, an average precision of 86.40%, and an average F1 Score of 87.78%.
5.2. SQL and NoSQL Database Experiments
These experiments performed six queries on different databases (Microsoft SQL Server, MongoDB, and Cassandra). The objective was to evaluate each query’s runtime, RAM used, and CPU percentage. We also evaluated databases scalability using a dataset with a double number of records. The dataset used in these experiments was the hospital data, consisting of Patients and Exams datasets. The tests were performed for each query three times, and the final result is the average of these three executions. It should be mentioned that there exists a reset between each run to eliminate cache effects, and therefore the three executions have similar results.
Two complex queries were created to extract valuable insights from the two tables, Patient (563,552 records) and CovidExam (935,960 records). The first query (Query Region—Query 1), in
Figure 9, returns different sets of sums, excluding the underline. There are fifth sums:
The first set is the total counts performed for COVID-19 (PCR and serological tests): how many of these tests were detected or reactive to COVID-19 and how many were not seen or not reactive COVID-19;
The second set is the total number of PCR tests: how many of these tests were detected by COVID-19, and how many were not seen by COVID-19;
The third set is the total number of serological tests: how many of these tests were reactive to COVID-19, and how many were non-reactive to COVID-19;
The fourth set is the total tests performed on male patients: how many of these tests were detected or reactive to COVID-19, and how many of these tests were not seen or not reactive to COVID-19;
Finally, the fifth set is the total tests performed on female patients: how many of these tests were not detected or not reactive to COVID-19.
All these sets are grouped by the Id of the patient’s region.
The second query (Query RegionYear—Query 2) in
Figure 9 includes the underline returns the same as the first query, but it is also grouped by the patient’s birth year.
Figure 10a,b show the runtime results for Query 1 and Query 2, respectively, during their execution in different databases. The scalability was assessed using ExamCovidPatient with 467,980 records (first test) and 935,960 records (second test). Analyzing the results, the performance of MongoDB is better than Microsoft SQL Server and better than Cassandra. In this case, we use one single table (ExamCovidPatient) to avoid the join operation on NoSQL databases.
For example, using Query 1, MongoDB presents the best results (1.76 s) compared to SQL Server (2.59 s) and Cassandra (7.14 s), using the smallest dataset. The runtime of SQL Server has an overhead of 1.47 times compared to MongoDB and Cassandra shows an even worse overhead of 4.05 times. This is because MongoDB is a distributed database by default, which allows horizontal scalability without any changes to application logic.
Analyzing the database scalability when the size increases two times, we expect a linear increase in runtime. However, all the databases show good scalability, presenting an increase in the runtime of only 1.99 (MongoDB), 1.49 (SQL Server), and 1.12 (Cassandra). In this case, we can conclude that all databases perform well when we increase the workload. Query 2 results are similar with only an insignificant increase in the runtime of all databases because of added complexity.
We also tested MongoDB with join queries, but it did not execute the queries in a reasonable amount of time. These tests show that MongoDB would take a very long time to execute the intended query, and further confirm that MongoDB is inadequate to join queries but performs very well in large datasets. Cassandra was connected to Apache Spark version 3.1.2. and it ran the join queries without any problems but with a significant execution time.
Figure 11a,b show the runtime results for Query 1 and Query 2, respectively, using SQL Server and Cassandra with joint operations. The runtime increases in both databases, but it is more noticeable in Cassandra because of mentioned weaknesses of NoSQL databases in these cases.
Figure 12a,b show the RAM usage for Query 1 and Query 2 during execution in different databases. Microsoft SQL Server used less memory than MongoDB and Cassandra in both queries. Using MongoDB, the difference is from a minimum of 101.6 MB to a maximum of 238.11 MB. This difference is marginal with Cassandra, where the minimum difference is 73.1, and the maximum is 111.7 MB when compared to SQL Server RAM use. This might be due to MongoDB and Cassandra’s use of data replication.
Figure 13a,b show the percentage of CPU usage results for Query 1 and Query 2, respectively, during its execution in the different databases. Microsoft SQL Server was observed to use less CPU than MongoDB and Cassandra when executing both queries with a maximum difference of 1.19× and 4.78×, respectively.
The third query (Query Orange—Query 3) was selected using an activated audit trail that controlled all the selects made in the Microsoft SQL Server database. When Orange Data Mining was connected to Microsoft SQL Server, and the Data Mining tests (classification tests) were performed, the audit trail controlled all queries that Orange needed to make in the database to perform the tests. Therefore, the third query evaluated was the query audit showed. This query, in
Figure 14, outputs the number of records in table
SymptomsCovid.
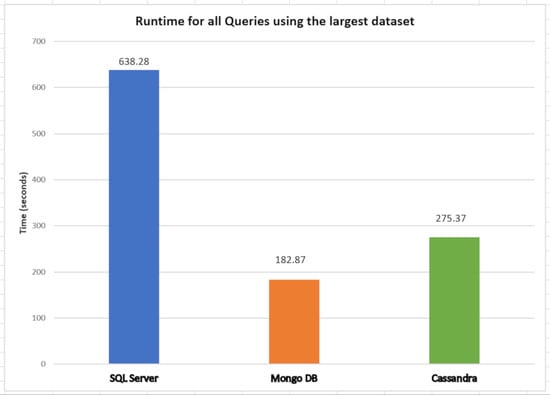
Figure 15a–c show the Query 3 results during its execution using the different databases. The scalability was assessed using SymptomsCovid with 1,510,722 records in the first test and 3,021,444 records in the second test. The results from
Figure 15a show that MongoDB and Cassandra were much better than Microsoft SQL Server in runtime as expected, since NoSQL databases rely on denormalization and try to optimize for the denormalized case. This factor makes queries much faster because data are stored in the same place (table or collection), and there is no need to perform a join. The relational model used by SQL Server proves to be quite harmful to queries in databases such as Query 3. It should be noted that SQL Server presents a maximum runtime overhead of 1341× compared to MongoDB (13.41/0.01).
In
Figure 15b, Microsoft SQL Server is shown to be the database that uses more memory for this query. Finally, according to
Figure 15c, none of the databases used much CPU (%) to execute this query, except Cassandra.
Considering the low number of records, a new table was created and named FactosExame (joint between Paciente table and Exame table), containing 16 fields and 26,651,928 records.
The fourth query (Query ExamFacts), in
Figure 16, is very similar to the first query (Query 1). The only difference is that there is no join because the information is gathered in one table containing all the hospital exams. The scalability was assessed using FactsExam (Exam plus Patient) with 13,325,964 records (first test) and 26,651,928 records (second test).
Figure 17a–c show the runtime, memory used, and CPU percentage used for the fourth query during its execution in the different databases.
The results from
Figure 17a show that SQL Server was the slowest to execute the query using the most extensive dataset (1.66× quiet than MongoDB and 1.26× than Cassandra). This is due to the limitations of SQL Server working with a large amount of data. On the other hand, MongoDB was faster than Cassandra by about 18 s (i.e., 1.31× faster).
In
Figure 17b, the results for the most extensive dataset show that SQL Server is the database that needed more memory to execute the query, followed by MongoDB with less than 1174.5 MB. Cassandra was by far the one that needed to use the least amount of memory, with a difference of 6504.9 MB for SQL Server and 5330.4 MB for MongoDB. SQL Server scales vertically, which means that increasing components such as RAM or CPU enable the increase of the load on a single machine. This explains the high memory values by SQL Server. MongoDB keeps all data in its caches, and it will not release them until it reaches the in-memory storage maximum threshold, which is by default 50% of physical RAM minus 1 GB. This means its memory will increase continuously when data are written or read. MongoDB will not free memory unless another process asks for it [
36]. This explains the high memory values used by MongoDB.
In
Figure 17c, SQL Server needs less CPU usage during the query execution for the largest dataset. Comparing the two NoSQL Databases, MongoDB used much less CPU percentage than Cassandra, separated by a difference of 4.28 times. The small CPU percentage obtained by MongoDB might be because MongoDB correctly made indexing.
Comparing the two NoSQL databases, it is possible to observe from the results in these last experiments that MongoDB was faster than Cassandra. MongoDB keeps all its data in cache memory, thus using more memory and consecutively using a lower CPU percentage. On the other hand, as Cassandra uses little memory, it does not keep its data in the cache, thus needing to use high values of CPU percentage to complete the execution of the query. For this reason, Cassandra is a bit slower when running the query.
We also evaluate the storage size of each database when storing the most extensive dataset, and the results are illustrated in
Figure 18. This figure shows that MongoDB was the best performing solution, followed by Cassandra and, lastly, Microsoft SQL Server (more 4.51× than MongoDB and 2.16× than Cassandra). MongoDB’s better performance is due to the WiredTiger storage engine that compresses data and indexes by default. Otherwise, the storage would be more significant.
We are interested in testing the databases with more complex queries that need more processing power. Therefore, we define two more queries: Query 5 and Query 6.
Query 5 is shown in
Figure 19 and returns the number of female and male patients who underwent COVID tests, which corresponds to only two rows.
Figure 20a–c show the runtime, memory used, and CPU percentage used for the fifth query using the different databases. The first test uses an Exam table with 13,325,964 records and the second one with 26,651,928 records.
Figure 20a shows the superior runtime of MongoDB: 4.16× better than SQL Server and 1.49× than Cassandra. Analyzing the scalability of each database when the number of records is duplicated, SQL Server shows an increment of 4.44×, MongoDB 1.56×, and Cassandra 1.49×. These results demonstrate the superior performance of NoSQL databases when dealing with large datasets.
The results for RAM and CPU usage are consistent with the previous test and show that Cassandra is the best in using RAM memory, and MongoDB uses less CPU.
Query 6 is shown in
Figure 21 and returns 21 rows with the federation unity of each state that underwent COVID tests. Each row has the number of female and male patients who underwent COVID tests, the number of different sample collection dates, the number of other sample collection locations, the number of different regions, the most present region, and the total counts performed for COVID-19 (PCR and serological tests), how many of these tests were detected or reactive to COVID-19 and how many were not detected or not reactive COVID-19. This is the most challenging query used in the experiments.
Figure 22a–c show the runtime, memory used, and CPU percentage used for the sixth query, where the first test uses an Exam table with 13,325,964 records and the second one 26,651,928 records.
Figure 22a shows better results than Query 5, with MongoDB achieving a runtime 4.75× better than SQL Server and 1.60× than Cassandra. Analyzing the scalability of each database when the number of records is duplicated, SQL Server shows an increment of 5.95×, MongoDB 1.77×, and Cassandra 1.68×.
The results for RAM and CPU usage are also consistent with the previous tests and show that Cassandra continues to be the best in the use of RAM and MongoDB in the utilization of CPU.
In summary, MongoDB and Cassandra are databases that do not require a schema, naturally making them more adaptable to changes.
Cassandra is a much more stationary database. It facilitates static typing and demands the categorization and definition of columns beforehand, requiring much less RAM. Additionally, the load characteristic of the application database to support also plays a crucial role. If we are expecting heavy load input, Cassandra will provide better results with its multiple master nodes. With stout load output, both MongoDB and Cassandra will show good performance.
Finally, many consider MongoDB or SQL Server to have the upper hand regarding consistency requirements. Ultimately, the decision between these three databases will depend on the application requirements and model.