1. Introduction
In the past decades, the use of herbicides, pesticides, and other chemical substances has continuously been increased. Unfortunately, overuse of these materials cause surface water pollution, environmental pollution, and animal and human toxicity [
1] (Liu and O’Connell, 2002). For this reason, scientists proposed the use of precision agriculture. One of the advantages of precision agriculture is the use of chemical substances only over the area of interest, called site-specific spray. The first step in site-specific spray operations is the proper recognition of the area of interest. Image processing is applied in this field to automatically identify plant species. In the past, there have been various researchers working on the recognition of different plants and trees.
Singh and Bhamrah [
2] stated that the recognition of different plants, especially medicine plants is challenging. Eight species of different plants including Amrood, Jaman, Kathal, Nimbu, Palak, Pippal, Sfaida and Toot, were used in this study. From each specimen, ten images were taken. The proposed algorithm had five main steps: image histogram equalization, pre-processing, segmentation, feature extraction, and classification. Seven features were extracted from each image to be used in the classifier. Extracted features were: solidity, length of the major axis, length of the minor axis, aspect ratio, area, perimeter, and eccentricity. The classifier used was an artificial neural network. Results showed that the accuracy of this classifier was 98.8%.
Kadir [
3] proposed a system to identify some leaves in different plants, based on the combination of Fourier descriptors and some shape features. Extracted features were: translation, scaling, moving the starting point (Fourier descriptors), convexity, irregularity factor, aspect factor, solidity, and roundness factor (shape features). To test the proposed algorithm, a total of 100 images were used. A Bayesian classifier was used. Results showed a classifier accuracy of 88%.
Ehsanirad [
4] classified several plant leaves based on texture features such as autocorrelation, entropy, contrast, correlation, and homogeneity. Two methods, gray level co-occurrence matrix (GLCM) and principal component analysis (PCA) were used to feature selection. Following image processing, a total of 65 images from different plants were used to validate and test the system. Results showed a classification accuracy with PCA and GLCM of 98% and 78%, respectively.
Mursalin et al. [
5], performed a study over the classification of five plant species, including Capsicum, Burcucumber, Cogongrass, Marsh herb, and Pigweed. To train and test the proposed system 400 images (80 images from each plant type) were used. Different features were extracted for classification purposes from each image. Extracted features were: solidity, elongatedness, convexity, form factor, area, thickness, perimeter, convex area, convex perimeter, and some color features. Three classifiers were used: support vector machine (SVM), C4.5 and naive Bayes. Results showed the best case of naive Bayes, SVM and C4.5 classifiers with accuracies of 99.3%, 98.24%, and 97.86%, respectively.
In Ahmed et al. [
6], authors classified six plants species including Capsicum frutescens L., Amaranthus viridis L., Enhydra fluctuans lour., Chenopodium album L., Imperata cylindrica (L.) P. beauv. and Sicyos angulatus L., based on the SVM classifier. In this study, a total of 224 images were taken from previously listed plants. In the next step, 14 features were extracted from each image in fields of color features, moment invariants, and size-independent shape features. Among previous features, the combination of only 9 features had the best accuracy results. Final nine features used were solidity, elongatedness, mean value component ‘r’ in normalized RGB color space, mean value ‘b’ in RGB color space, standard deviation ‘r’ component, standard deviation of ‘b’ component in RGB color space, the
, the
, and the
, of area them all. Results showed that the SVM classifier had an accuracy of 97.3%.
Rumpf et al. [
7], used three sequential SVM to classify 10 different plant types including Agropyron repens, Alopecurus Myosuroides, C. Arvense, Galium aparine, Lamium sp., Matricaria inodora, Sinapis arvensis, Stellaria media, Veronica persica, and Hordeum vulgare. From each image, 10 features were extracted: number of pixels of an object, mean distance of the border to the center of gravity, maximum distance of the border to the center of gravity, vertical distance of the border to the main axis of the object, eccentricity, first two-moment invariants, skelmean, and skelsize. Results showed that total accuracy classification for the first SVM classifier was 97.7%, and for the second and third 80%.
Pereira et al. [
8], classified three different plant types based on shape features. These plants were:
E. crassipes, P. stratiotes, and
S. auriculata. In order to train and test the proposed system, a total of 51 images of
E. crassipes, 46 images of
P. stratiotes and 63 images of
S. auriculata were taken. Several features including beam angle statistics, FD, moment invariants, multiscale fractal dimension, and tensor scale descriptor, were extracted. Results showed that a neural network classified these three plant types with an accuracy above 80%.
In addition, Azlah et al. [
9], have recently presented a thorough review paper about plant leaf recognition and classification techniques, concluding that the current image processing techniques should be robust under diverse intensity lighting conditions, which could, in turn, be developed by tweaking the detection technique leading to detection of specific plant diseases.
The aim of this study is to design an intelligent computer vision system to classify five species tree leaves, including 1–
Cydonia oblonga (quince), 2–
Eucalyptus camaldulensis dehn (river red gum), 3–
Malus pumila (apple), 4–
Pistacia atlantica (mt. Atlas mastic tree) and 5–
Prunus armeniaca (apricot).
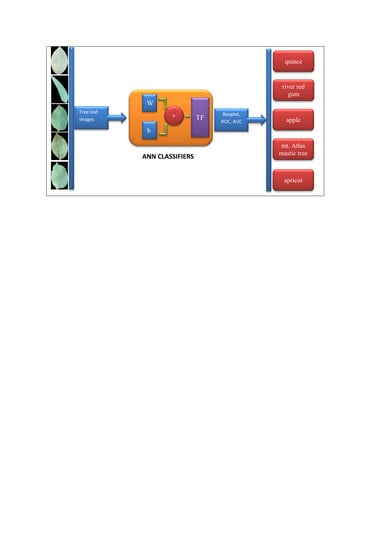
Figure 1 depicts a flowchart of the computer vision system for plant tree leaves automatic classification, here proposed.
2. Materials and Methods
As it can be seen from
Figure 1, the proposed computer vision system consists of five steps: the first step includes imaging, the second step includes segmentation and pre-processing, the third step includes feature extraction (including texture descriptors based on the histogram, texture features based on the gray level co-occurrence matrix, shape features, moment invariants, and color features), fourth step includes discriminant feature selection (based on two methods: hybrid artificial neural network–differential evolution (ANN–DE) and hybrid artificial neural network–genetic algorithm (ANN–GA)), and fifth step comprises classification based on three different classifiers, including hybrid artificial neural network–ant bee colony (ANN–ABC), hybrid artificial neural network–biogeography based optimization (ANN–BBO) and Fisher’s linear discriminant analysis (LDA). Each of the five steps in the here proposed computer vision system, depicted in
Figure 1 flowchart, will be further explained with more detail in the next paper sections.
2.1. Database Used in Computer Vision
As already mentioned, in this study five different types of leaves were investigated, including 1—
Cydonia oblonga (quince), see
Appendix Figures A1 and A2—
Eucalyptus camaldulensis dehn (river red gum), see
Appendix Figures A2 and A3—
Malus pumila (apple), see
Appendix Figures A3 and A4—
Pistacia atlantica (mt. Atlas mastic tree), see
Appendix Figures A4 and A5—
Prunus armeniaca (apricot), see
Appendix Figure A5.
Table 1 shows the class number, English common tree name, scientific tree name and the number of samples in each class.
Figure 2 shows one sample of each leaf type. All images were taken in Kermanshah, Iran (longitude: 7.03°E; latitude: 4.22°N). High-quality images were taken with a color GigE industrial camera (ImagingSource, model DFK-23GM021, 1/3 inch Aptina CMOS MT9M021 sensor, 1.2 MP 1280 × 960 spatial resolution, Germany), mounting an appropriate Computar CBC Group lens (model H0514-MP2, f = 5 mm F1.4, 1/2 inch type megapixel cameras, Japan). The camera was fixed at 10 cm above ground level, and all images were taken under white light of 327 lux lighting conditions.
2.2. Image Pre-Processing and Segmentation
The segmentation stage is an important one in image processing, since in case of wrong segmentation, either the background is considered as an object (in this study, any tree leaf) or object is considered as background. To reach a proper segmentation, six standard color spaces (RGB, HSV, HSI, YIQ, CMY, and YCbCr) were taken into account to find the best color space for segmentation purposes. Results have shown that the best color space for image segmentation was YCbCr color space and the best channels for image thresholding were luminance Y and chrominance Cr channels. Equation (1) was used to set the value of the threshold used to segment objects from their background in tree leaf images:
The implications of this equation are as follows: each pixel in YCbCr color space that has Y components smaller or equal to 100, or Cr components larger or equal to 15, is considered as background, otherwise, a pixel is considered as foreground (tree leaf object). In order to extract shape features from segmented images, binary images are needed. To extract shape features with high accuracy, some morphological operations are needed, since usually, some “noisy” pixels can exist in segmented images. For this reason, the Matlab imclose function was used, (Gonzalez et al. [
10]).
Figure 3 summarizes pre-processing operations, shown over three sample tree leaf images.
2.3. Feature Extraction
It is well-known that in order to properly classify different tree leaf types, feature extraction is needed. Features of various types and in different fields include texture descriptors based on the histogram, texture features based on the gray level co-occurrence matrix, shape features, moment invariants and color features (others may exist that are not used here). Above mentioned feature types were extracted from each leave. Indeed, the first 285 features of each leaf image were extracted and then effective (high discriminant) features were selected among them by hybrid ANN–DE and hybrid ANN–GA optimization approaches.
2.3.1. Texture Descriptors Based on the Histogram
Texture features that were extracted based on histogram include homogeneity, entropy, smoothness, third moment, average and standard deviation.
2.3.2. Texture Features Based on the Gray Level Co-Occurrence Matrix (GLCM)
It is well-known that neighbor angles have a big impact on the estimation of the values of each feature based on the GLCM.
Table 2 shows texture features extracted based on the gray level co-occurrence matrix. These features were extracted for four different angles: 0°, 45°, 90°, and 135°. Thus, in total, 27 × 4 = 108 texture features were extracted from each object.
2.3.3. Shape Features
In this study, 29 shape features were extracted.
Table 3 lists the shape features that were used here.
2.3.4. Moment Invariant Features
Moment invariant features have the advantage that are insensitive to translation, reflection dilation, and rotation. In this study, 10 moment-invariant features were extracted: first-order moment invariant, second-order moment invariant, third-order moment invariant, fourth-order moment invariant, fifth-order moment invariant, sixth-order moment invariant, seventh order moment invariant, difference of first and seventh order moment invariants, difference of second and sixth-order moment invariants and difference of third and fifth-order moment invariants.
2.3.5. Color Features
Different color features have different values in different color spaces. For this reason, some color features were extracted in RGB, YCbCr, YIQ, CMY, HSV and HSI color spaces. Extracted color features are divided into two groups:
‘Statistical’ Color Features
These features comprised mean and standard deviations of the first color component, the second component, third component and mean of first, second and third components for all RGB, YCbCr, YIQ, CMY, HSV, and HSI color spaces. Thus, the total number of ‘statistical’ color features was .
Vegetation Index Color Features
Table 4 lists 14 vegetation index features for RGB color space, including mathematical definitions. These features were also computed for the other five color spaces (YCbCr, YIQ, CMY, HSV, and HSI) totaling
vegetation index features.
2.4. Discriminant Feature Selection
As mentioned before, 285 features were extracted from each leaf image object. The use of all features as input to classifiers is not wise, given the problem of overfitting and poor generalization to the test set. The selection of discriminant groups of features is a well-known good practice. To do so, two methods based on artificial intelligence were used to select effective features (results to be shown later on):
2.5. Optimal Neural Network Classifier Parameters
Classification is the final step in designing a computer or machine vision system. A high-performance classifier guarantees the accuracy of the computer version system. In this study, three classifiers including hybrid ANN–ABC, hybrid ANN–BBO and Fisher’s linear discriminant analysis (LDA), were used in classifying. In order to have statistically valid results, 100 uniform random simulations were repeated. All data were divided into two disjoint groups, training, and validation set data (60% input samples) and test set data (40% of input samples) in each of the 100 averaged simulations, following a uniform random distribution with probability 0.6 and 0.4 to belong to train/validation and test sets, respectively. It is worth mention that a multilayer perceptron (MLP) ANN was used. An MLP neural network has five adjustable parameters: number of neural network layers, number of neurons in each layer, nonlinear transfer function, back-propagation network training function, and back-propagation weight/bias learning function. ABC and BBO algorithms were used to select optimal MLP parameter values.
Table 5 shows the optimum values of the MLP parameters which were determined with ABC and BBO algorithms.