Detection of Adulteration in Infant Formula Based on Ensemble Convolutional Neural Network and Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. NIR Spectroscopy Measurement
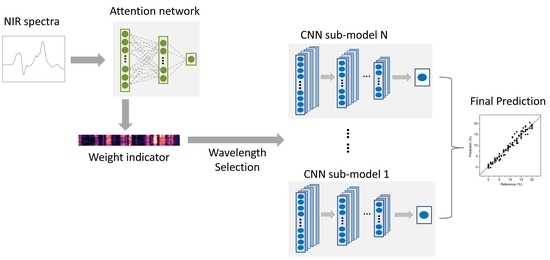
2.3. Wavelength Selection Based on Attention Mechanism
2.4. Convolutional Neural Network
2.5. Architecture of the AM-ECNN
2.6. Methods for Comparison
2.7. Data Processing, Model Optimization and Evaluation
3. Results and Discussion
3.1. PCA Analysis
3.2. Attention Mechanism Based on the Weight Indicator
3.3. Parameter Optimization
3.4. Results Comparison
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tyan, Y.C.; Yang, M.H.; Jong, S.B.; Wang, C.K.; Shiea, J. Melamine contamination. Anal. Bioanal. Chem. 2009, 395, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.M.; Pereira-Filho, E.R.; Rodriguez-Saona, L.E. Rapid detection and quantification of milk adulteration using infrared microspectroscopy and chemometrics analysis. Food Chem. 2013, 138, 19–24. [Google Scholar] [CrossRef]
- Das, S.; Sivaramakrishna, M.; Biswas, K.; Goswami, B. Performance study of a ‘constant phase angle based’ impedance sensor to detect milk adulteration. Sens. Actuat. A-Phys. 2011, 167, 273–278. [Google Scholar] [CrossRef]
- Hirwitz, D.; Latimer, G. Official Methods of Analysis. In Aoac International, 16th ed.; Sec. 33.2.11, Method 991.20 (2); A.O.A.C. International: Gaithersburg, MD, USA, 1995. [Google Scholar]
- Rowland, S.J. The determination of the nitrogen distribution in milk. J. Dairy Res. 1937, 9, 42–46. [Google Scholar] [CrossRef]
- Sáez-Plaza, P.; Michałowski, T.; Navas, M.J.; Asuero, A.G.; Wybraniec, S. An Overview of the Kjeldahl Method of Nitrogen Determination. Part, I. Early History, Chemistry of the Procedure, and Titrimetric Finish. Crit. Rev. Anal. Chem. 2013, 43, 178–223. [Google Scholar] [CrossRef]
- Venkatasami, G.; Sowa, J.R. A rapid, acetonitrile-free, HPLC method for determination of melamine in infant formula. Anal. Chim. Acta 2010, 665, 227–230. [Google Scholar] [CrossRef]
- Miao, H.; Fan, S.; Wu, Y.N.; Zhang, L.; Zhou, P.P.; Chen, H.J.; Zhao, Y.F.; Li, J.G. Simultaneous determination of melamine, ammelide, ammeline, and cyanuric acid in milk and milk products by gas chromatography-tandem mass spectrometry. Biomed. Environ. Sci. 2009, 02, 87–94. [Google Scholar] [CrossRef]
- Múller, L.; Barták, P.; Bednář, P.; Fryšová, I.; Ševčík, J.; Lemr, K. Capillary electrophoresis-mass spectrometry—A fast and reliable tool for the monitoring of milk adulteration. Electrophoresis 2010, 29, 2088–2093. [Google Scholar] [CrossRef]
- Dong, Y.L.; Yan, N.; Li, X.; Zhou, X.M.; Zhou, L.; Zhang, H.J.; Chen, X.G. Rapid and sensitive determination of hydroxyproline in dairy products using micellar electrokinetic chromatography with laser-induced fluorescence detection. J. Chromatogr. A 2012, 1233, 156–160. [Google Scholar] [CrossRef]
- Lei, H.; Shen, Y.; Song, L.; Yang, J.; Chevallier, O.P.; Haughey, S.A.; Wang, H.; Sun, Y.; Elliott, C.T. Hapten synthesis and antibody production for the development of a melamine immunoassay. Anal. Chim. Acta 2010, 665, 84–90. [Google Scholar] [CrossRef]
- Bania, J.; Ugorski, M.; Polanowski, A. Application of polymerase chain reaction for detection of goats’ milk adulteration by milk of cow. J. Dairy Res. 2001, 68, 333. [Google Scholar] [CrossRef] [PubMed]
- Fodey, T.L.; Thompson, C.S.; Traynor, I.M.; Traynor, I.M.; Haughey, S.A.; Kennedy, D.G.; Crooks, S.R.H. Development of an optical biosensor based immunoassay to screen infant formula milk samples for adulteration with melamine. Anal. Chem. 2011, 83, 5012–5016. [Google Scholar] [CrossRef] [PubMed]
- Brandao, M.P.; Neto, M.G.; Anjos, V.C.; Bell, M.J.V. Detection of adulteration of goat milk powder with bovine milk powder by front-face and time resolved fluorescence. Food Control 2017, 81, 168–172. [Google Scholar] [CrossRef]
- Mabood, F.; Jabeen, F.; Hussain, J.; Al-Harrasi, A.; Hamaed, A.; Al Mashaykhi, S.A.A.; Al Rubaiey, Z.M.A.; Manzoor, S.; Khan, A.; Haq, Q.M.I.; et al. FT-NIRS coupled with chemometric methods as a rapid alternative tool for the detection & quantification of cow milk adulteration in camel milk samples. Vib. Spectrosc. 2017, 92, 245–250. [Google Scholar]
- Capuano, E.; Boerrigter-Eenling, R.; Koot, A.; Ruth, S.M. Targeted and untargeted detection of skim milk powder adulteration by near-infrared spectroscopy. Food Anal. Methods 2015, 8, 2125–2134. [Google Scholar] [CrossRef]
- Botros, L.L.; Jablonski, J.; Chang, C.; Bergana, M.M.; Wehling, P.; Harnly, J.M.; Downey, G.; Harrington, P.; Potts, A.R.; Moore, J.C. Exploring authentic skim and nonfat dry milk powder variance for the development of nontargeted adulterant detection methods using near-infrared spectroscopy and chemometrics. J. Agric. Food Chem. 2013, 61, 9810–9818. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.V.S.; Fernandes, D.D.S.; Araújo, M.C.U.; Diniz, P.H.G.D.; Maciel, M.I.S. Simultaneous determination of goat milk adulteration with cow milk and their fat and protein contents using NIR spectroscopy and PLS algorithms. LWT Food Sci. Technol. 2020, 127, 109427. [Google Scholar] [CrossRef]
- Zhang, L.G.; Zhang, X.; Ni, L.J.; Xue, Z.B.; Gu, X.; Huang, S.X. Rapid identification of adulterated cow milk by non-linear pattern recognition methods based on near infrared spectroscopy. Food Chem. 2014, 145, 342–348. [Google Scholar] [CrossRef]
- Harnly, J.M.; Harrington, P.B.; Botros, L.L.; Jablonski, J.; Chang, C.; Bergana, M.M.; Wehling, P.; Downey, G.; Potts, A.R.; Moore, J.C. Characterization of Near-infrared spectral variance in the authentication of skim and nonfat dry milk powder collection using ANOVA-PCA, pooled-ANOVA, and partial least-squares regression. J. Agric. Food Chem. 2014, 62, 8060–8067. [Google Scholar] [CrossRef]
- Borin, A.; Ferrão, M.F.; Mello, C.; Maretto, D.A.; Poppi, R.J. Least-squares support vector machines and near infrared spectroscopy for quantification of common adulterants in powdered milk. Anal. Chim. Acta 2006, 579, 25–32. [Google Scholar] [CrossRef]
- Balabin, R.M.; Smirnov, S.V. Melamine detection by mid- and near-infrared (MIR/NIR) spectroscopy: A quick and sensitive method for dairy products analysis including liquid milk, infant formula, and milk powder. Talanta 2011, 85, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Mabood, F.; Jabeen, F.; Ahmed, M.; Hussain, J.; Al Mashaykhi, S.A.A.; Al Rubaiey, Z.M.A.; Farooq, S.; Boqué, R.; Ali, L.; Hussain, Z.; et al. Development of new NIR-spectroscopy method combined with multivariate analysis for detection of adulteration in camel milk with goat milk. Food Chem. 2017, 221, 746–750. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L. Gradient-based learning applied to document recognition. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Lussier, F.; Missirlis, D.; Spatz, J.P.; Masson, J.F. Machine-Learning-Driven Surface-Enhanced Raman Scattering Optophysiology Reveals Multiplexed Metabolite Gradients near Cells. ACS Nano 2019, 13, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Kim, H. Intact metabolite spectrum mining by deep learning in proton magnetic resonance spectroscopy of the brain. Magn. Reson. Med. 2019, 82, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Aljakouch, K.; Hilal, Z.; Daho, I.; Schuler, M.; Krauß, S.D.; Yosef, H.K.; Dierks, J.; Mosig, A.; Gerwert, K.; El-Mashtoly, S.F. Fast and Noninvasive Diagnosis of Cervical Cancer by Coherent Anti-Stokes Raman Scattering. Anal. Chem. 2019, 91, 13900–13906. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Jenkins, C.; Shermer, S.M.; Langbein, F.C. MRSNet: Metabolite quantification from edited magnetic resonance spectra with convolutional neural networks. arXiv 2019, arXiv:1909.03836. [Google Scholar]
- Acquarelli, J.; Laarhoven, T.; Gerretzen, J.; Thanh, N.T.; Lutgarde, M.C.B.; Elena, M. Convolutional neural networks for vibrational spectroscopic data analysis. Anal. Chim. Acta 2017, 954, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Cui, C.H.; Fearn, T. Modern practical convolutional neural networks for multivariate regression: Applications to NIR calibration. Chemometr. Intell. Lab. 2018, 182, 9–20. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wang, Z.B. Feature selection based convolutional neural network pruning and its application in calibration modeling for NIR spectroscopy. Chemometr. Intell. Lab. 2019, 191, 103–108. [Google Scholar] [CrossRef]
- He, H.; Xu, M.X.; Zong, C.; Zheng, P.; Luo, L.L.; Wang, L.; Ren, B. Speeding up the line-scan Raman imaging of living cells by deep convolutional neural network. Anal. Chem. 2019, 91, 7070–7077. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.D.; Rivenson, Y.; Jin, Y.Y.; Wei, Z.S.; Gao, R.; Günaydın, H.; Bentolila, L.A.; Kural, C.; Ozcan, A. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 2019, 16, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, A.; Tsentalovich, Y.P.; Yanshole, V.V. Deep learning for the precise peak detection in high-resolution LC-MS data. Anal. Chem. 2020, 92, 588–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, N.; Hattori, Y.; Nagata, T.; Shinya, S.; Güntert, P.; Kojima, C.; Fujiwara, T. Noise peak filtering in multi-dimensional NMR spectra using convolutional neural networks. Bioinformatics 2018, 34, 4300–4301. [Google Scholar] [CrossRef] [PubMed]
- Krauß, S.D.; Roy, R.; Yosef, H.K.; Lechtonen, T.; El-Mashtoly, S.F.; Gerwert, K.; Mosig, A. Hierarchical deep convolutional neural networks combine spectral and spatial information for highly accurate Raman-microscopy-based cytopathology. J. Biophotonics 2018, 11, e201800022. [Google Scholar] [CrossRef]
- Liu, J.; Osadchy, M.; Ashton, L.; Foster, M.; Solomon, C.J.; Gibson, S.J. Deep convolutional neural networks for Raman spectrum recognition: A unified solution. Analyst 2017, 142, 4067–4074. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Tan, C.; Lin, Z. Ensemble of extreme learning machines for multivariate calibration of near-infrared spectroscopy. Spectrochim. Acta A 2020, 229, 117982. [Google Scholar] [CrossRef]
- Tan, C.; Li, M.; Qin, X. Random Subspace Regression Ensemble for Near-Infrared Spectroscopic Calibration of Tobacco Samples. Anal. Sci. Int. J. Jpn. Soc. Anal. Chem. 2008, 24, 647–653. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.; Wang, J.; Qin, X.; Li, M. Ensemble multivariate calibration based on mutual information for food analysis using near-infrared spectroscopy. Anal. Lett. 2010, 43, 2640–2651. [Google Scholar] [CrossRef]
- Tan, C.; Wang, J.; Wu, T.; Qin, X.; Li, M. Determination of nicotine in tobacco samples by near-infrared spectroscopy and boosting partial least squares. Vib. Spectrosc. 2010, 54, 35–41. [Google Scholar] [CrossRef]
- Bian, X.; Li, S.; Shao, X.; Liu, P. Variable space boosting partial least squares for multivariate calibration of near-infrared spectroscopy. Chemometr. Intell. Lab. 2016, 158, 174–179. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, Y.P.; Tang, L.J.; Wu, H.L.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Ensemble preprocessing of near-infrared (NIR) spectra for multivariate calibration. Anal. Chim. Acta 2008, 616, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Li, Y.; Wu, Z.; Zhang, Q.; Zheng, Z.; Shi, X.; Qiao, Y. A online NIR sensor for the pilot-scale extraction process in fructus aurantii coupled with single and ensemble methods. Sensors 2015, 15, 8749–8763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, X.; Bian, X.; Cai, W. An improved boosting partial least squares method for near-infrared spectroscopic quantitative analysis. Anal. Chim. Acta 2010, 666, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Tsakiridis, N.L.; Tziolas, N.V.; Theocharis, J.B.; Zalidis, G.C. A genetic algorithm-based stacking algorithm for predicting soil organic matter from vis-NIR spectral data. Eur. J. Soil Sci. 2019, 70, 578–590. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, Y.; Zhang, Q.; Shi, X.; Wu, Z.; Qiao, Y. Comparison of ensemble strategies in online NIR for monitoring the extraction process of pericarpium citri reticulatae based on different variable selections. Planta Med. 2016, 82, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Bi, Y.; Xie, Q.; Peng, S.; Tang, L.; Hu, Y.; Tan, J.; Zhao, Y.; Li, C. Dual stacked partial least squares for analysis of near-infrared spectra. Anal. Chim. Acta 2013, 792, 19–27. [Google Scholar] [CrossRef]
- Bian, X.; Zhang, C.; Tan, X.; Dymek, M.; Guo, Y.; Lin, L.; Cheng, B.; Hu, X. Boosting extreme learning machine for near-infrared spectral quantitative analysis of diesel fuel and edible blend oil samples. Anal. Methods 2017, 9, 2983–2989. [Google Scholar] [CrossRef]
- Chakraborty, D.; Saha, S.; Ghoshal, S. NIR spectrometry-based milk fat content classification using bagging ensembles. In Computational Advancement in Communication Circuits and Systems 2015; Springer: New Delhi, India, 2015; Volume 335, pp. 491–497. [Google Scholar]
- Merkwirth, C.; Mauser, H.; Gasch, T.S.; Roche, O.; Stahl, M.; Lengauer, T. Ensemble methods for classification in cheminformatics. J. Chem. Inf. Comput. Sci. 2004, 44, 1971–1978. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Guan, J.; Zhu, Q. Predicting bruise susceptibility in apples using Vis/SWNIR technique combined with ensemble learning. Int. J. Agric. Biol. Eng. 2017, 10, 144–153. [Google Scholar]
- Liu, Y.S.; Zhou, S.B.; Han, W.; Liu, W.X.; Qiu, Z.F.; Li, C. Convolutional neural network for hyperspectral data analysis and effective wavelengths selection. Anal. Chim. Acta 2019, 1086, 46–54. [Google Scholar] [CrossRef]
- Liu, Y.S.; Zhou, S.B. Rapid detection of hydrolyzed leather protein adulteration in infant formula by near-infrared spectroscopy. Food Sci. Technol. Res. 2017, 23, 469–474. [Google Scholar] [CrossRef] [Green Version]
- Maas, A.L.; Hannun, A.Y.; Ng, A.Y. Rectifier nonlinearities improve neural network acoustic models. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 17–19 June 2013; Volume 28, p. 3. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 7–9 July 2015. [Google Scholar]
- Wold, S.; Sjstrm, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemometr. Intell. Lab. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Chen, H.; Lin, B.; Cai, K.; Chen, A.; Hong, S. Quantitative analysis of organic acids in pomelo fruit using FT-NIR spectroscopy coupled with network kernel PLS regression. Infrared Phys. Technol. 2021, 112, 103582. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by random Forest. R News 2002, 2, 18–22. [Google Scholar]







| Layers | Number of Filters | Kernel Size | Stride | Padding | Nonlinear Activation |
|---|---|---|---|---|---|
| Convolution Layer 1 | 16 | 5 | 1 | Yes | LeakyReLU |
| Convolution Layer 2 | 16 | 5 | 1 | Yes | LeakyReLU |
| Convolution Layer 3 | 16 | 5 | 1 | Yes | LeakyReLU |
| Output | / | / | / | / | Sigmoid |
| RMSEP | R2 | RPD | |
|---|---|---|---|
| PLS 1 | 1.600 ± 0.295 | 0.930 ± 0.025 | 4.157 ± 0.728 |
| Kernel PLS 2 | 1.444 ± 0.254 | 0.949 ± 0.021 | 4.933 ± 1.085 |
| CNN 3 | 1.546 ± 0.366 | 0.933 ± 0.029 | 4.412 ± 1.197 |
| RF 4 | 1.780 ± 0.393 | 0.911 ± 0.042 | 3.789 ± 0.813 |
| RC-ECNN 5 | 1.225 ± 0.238 | 0.957 ± 0.019 | 5.548 ± 1.516 |
| AE-ECNN 6 | 1.168 ± 0.231 | 0.961 ± 0.016 | 5.804 ± 1.818 |
| RMSEP | R2 | RPD | |
|---|---|---|---|
| PLS 1 | 0.263 ± 0.058 | 0.987 ± 0.010 | 11.138 ± 4.052 |
| Kernel PLS 2 | 0.250 ± 0.044 | 0.992 ± 0.008 | 12.377 ± 4.826 |
| CNN 3 | 0.200 ± 0.041 | 0.992 ± 0.005 | 14.973 ± 6.975 |
| RF 4 | 0.349 ± 0.093 | 0.980 ± 0.011 | 8.244 ± 2.235 |
| RC-ECNN 5 | 0.164 ± 0.032 | 0.995 ± 0.004 | 17.811 ± 6.228 |
| AE-ECNN 6 | 0.159 ± 0.028 | 0.995 ± 0.004 | 18.004 ± 5.662 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhou, S.; Han, W.; Li, C.; Liu, W.; Qiu, Z.; Chen, H. Detection of Adulteration in Infant Formula Based on Ensemble Convolutional Neural Network and Near-Infrared Spectroscopy. Foods 2021, 10, 785. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10040785
Liu Y, Zhou S, Han W, Li C, Liu W, Qiu Z, Chen H. Detection of Adulteration in Infant Formula Based on Ensemble Convolutional Neural Network and Near-Infrared Spectroscopy. Foods. 2021; 10(4):785. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10040785
Chicago/Turabian StyleLiu, Yisen, Songbin Zhou, Wei Han, Chang Li, Weixin Liu, Zefan Qiu, and Hong Chen. 2021. "Detection of Adulteration in Infant Formula Based on Ensemble Convolutional Neural Network and Near-Infrared Spectroscopy" Foods 10, no. 4: 785. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10040785