Interaction Mechanism of Flavonoids and α-Glucosidase: Experimental and Molecular Modelling Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. α-Glucosidase Inhibitory Assay
2.3. Determination of Interaction Behaviors of Myricetin and Dihydromyricetin
2.4. Molecular Docking Method
2.5. IGM Method
2.6. Statistical Analysis
3. Results and Discussion
3.1. α-Glucosidase Inhibitory Activities
3.2. Fluorescence Analysis
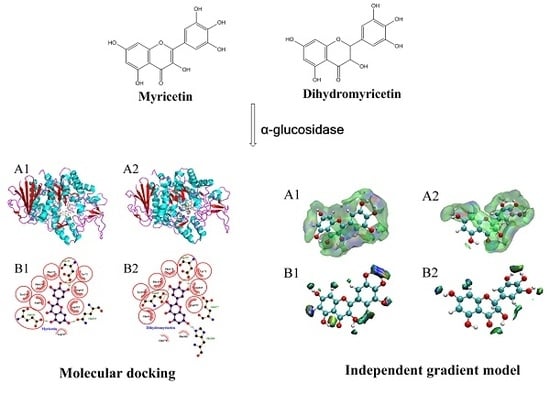
3.3. Molecular Docking Analysis
3.4. IGM Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, L.; Deseo, M.A.; Morris, C.; Winter, K.M.; Leach, D.N. Investigation of α-glucosidase inhibitory activity of wheat bran and germ. Food Chem. 2011, 126, 553–561. [Google Scholar] [CrossRef]
- Zimmet, P.; Alberti, K.; Shaw, J. Global and societal implications of the diabetes epidemic. Nature 2001, 414, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Ghani, U. Re-exploring promising α-glucosidase inhibitors for potential development into oral anti-diabetic drugs: Finding needle in the haystack. Eur. J. Med. Chem. 2015, 103, 133–162. [Google Scholar] [CrossRef] [PubMed]
- Alongi, M.; Anese, M. Effect of coffee roasting on in vitro α-glucosidase activity: Inhibition and mechanism of action. Food Res. Int. 2018, 111, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Kadouh, H.C.; Sun, S.; Zhu, W.; Zhou, K. α-Glucosidase inhibiting activity and bioactive compounds of six red wine grape pomace extracts. J. Funct. Foods 2016, 26, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Erlund, I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Wang, T.Y.; Li, Q.; Bi, K.S. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef]
- Yang, B.; Liu, H.; Yang, J.; Gupta, V.K.; Jiang, Y. New insights on bioactivities and biosynthesis of flavonoid glycosides. Trends Food Sci. Technol. 2018, 79, 116–124. [Google Scholar] [CrossRef]
- Ong, K.C.; Khoo, H.E. Effects of myricetin on glycemia and glycogen metabolism in diabetic rats. Life Sci. 2000, 67, 1695–1705. [Google Scholar] [CrossRef]
- Shi, L.Y.; Zhang, T.; Liang, X.Y.; Hu, Q.; Huang, J.; Zhou, Y.; Chen, M.L.; Zhang, Q.Y.; Zhu, J.D.; Mi, M.T. Dihydromyricetin improves skeletal muscle insulin resistance by inducing autophagy via the AMPK signaling pathway. Mol. Cell. Endocrinol. 2015, 409, 92–102. [Google Scholar] [CrossRef]
- Li, Y.Q.; Zhou, F.C.; Gao, F.; Bian, J.S.; Shan, F. Comparative evaluation of quercetin, isoquercetin and rutin as inhibitors of α-glucosidase. J. Agric. Food Chem. 2009, 57, 11463–11468. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhang, G.; Liao, Y.; Gong, D. Inhibitory kinetics and mechanism of kaempferol on α-glucosidase. Food Chem. 2016, 190, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Proença, C.; Freitas, M.; Ribeiro, D.; Oliveira, E.F.; Sousa, J.L.; Tomé, S.M.; Ramos, M.J.; Silva, A.M.; Fernandes, P.A.; Fernandes, E. α-Glucosidase inhibition by flavonoids: An in vitro and in silico structure–activity relationship study. J. Enzym. Inhib. Med. Chem. 2017, 32, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Sarian, M.N.; Ahmed, Q.U.; So’ad, M.; Zaiton, S.; Alhassan, A.M.; Murugesu, S.; Perumal, V.; Mohamad, S.; Akilah, S.N.; Khatib, A. Antioxidant and antidiabetic effects of flavonoids: A structure-activity relationship based study. BioMed Res. Int. 2017, 2017, 8386065. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.W.; Li, X.; Sun, W.L.; Xing, Y.; Xiu, Z.L.; Zhuang, C.L.; Dong, Y.S. Dietary flavonoids and acarbose synergistically inhibit α-glucosidase and lower postprandial blood glucose. J. Agric. Food Chem. 2017, 65, 8319–8330. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, F.; Li, P.; She, Y.; Gao, W. Investigation of the interaction for three Citrus flavonoids and α-amylase by surface plasmon resonance. Food Res. Int. 2017, 97, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi-Kaji, S.; Shareghi, B.; Saboury, A.A.; Farhadian, S. Spectroscopic and molecular docking studies on the interaction between spermidine and pancreatic elastase. Int. J. Biol. Macromol. 2019, 131, 473–483. [Google Scholar] [CrossRef]
- Trivella, D.B.; Dos Reis, C.V.; Lima, L.M.T.; Foguel, D.; Polikarpov, I. Flavonoid interactions with human transthyretin: Combined structural and thermodynamic analysis. J. Struct. Biol. 2012, 180, 143–153. [Google Scholar] [CrossRef]
- Zeng, H.J.; Wang, Y.P.; Yang, R.; You, J.; Qu, L.B. Inhibitory effects of daidzein and genistein on trypsin: Insights from spectroscopic and molecular docking studies. Int. J. Biol. Macromol. 2016, 89, 336–343. [Google Scholar] [CrossRef]
- Li, A.; Maurel, F.; Delamar, M.; Wang, B. ONIOM study of the nonbonding interaction of the 2PU inhibitor with the CDK2 and CDK4 cyclin-dependant kinases. Int. J. Quantum Chem. 2009, 109, 1148–1157. [Google Scholar] [CrossRef]
- Lefebvre, C.; Rubez, G.; Khartabil, H.; Boisson, J.-C.; Contreras-García, J.; Hénon, E. Accurately extracting the signature of intermolecular interactions present in the NCI plot of the reduced density gradient versus electron density. Phys. Chem. Chem. Phys. 2017, 19, 17928–17936. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Liu, B.; Mo, H.; Liang, G. Comparative evaluation of tannic acid inhibiting α-glucosidase and trypsin. Food Res. Int. 2015, 76, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J. Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements. J. Mol. Model. 2007, 13, 1173–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Xu, D.; Wang, Q.; Zhang, W.; Hu, B.; Zhou, L.; Zeng, X.; Sun, Y. Inhibitory activities of caffeoylquinic acid derivatives from Ilex kudingcha CJ Tseng on α-glucosidase from Saccharomyces cerevisiae. J. Agric. Food Chem. 2015, 63, 3694–3703. [Google Scholar] [CrossRef] [PubMed]
- Dolatabadi, J.E.N.; Panahi-Azar, V.; Barzegar, A.; Jamali, A.A.; Kheirdoosh, F.; Kashanian, S.; Omidi, Y. Spectroscopic and molecular modeling studies of human serum albumin interaction with propyl gallate. RSC Adv. 2014, 4, 64559–64564. [Google Scholar] [CrossRef]
- Androutsopoulos, V.P.; Papakyriakou, A.; Vourloumis, D.; Spandidos, D.A. Comparative CYP1A1 and CYP1B1 substrate and inhibitor profile of dietary flavonoids. Bioorg. Med. Chem. 2011, 19, 2842–2849. [Google Scholar] [CrossRef] [PubMed]
- Mahfoudi, R.; Djeridane, A.; Benarous, K.; Gaydou, E.M.; Yousfi, M. Structure-activity relationships and molecular docking of thirteen synthesized flavonoids as horseradish peroxidase inhibitors. Bioorg. Chem. 2017, 74, 201–211. [Google Scholar] [CrossRef] [PubMed]




 | ||||||||||||
| No. | Type | Name | 3 | 4 | 5 | 6 | 7 | 2′ | 3′ | 4′ | 5′ | IC50 |
| 1 | I | myricetin | OH | OH | OH | OH | OH | OH | 2.09 | |||
| 2 | I | fisetin | OH | OH | OH | OH | 5.50 | |||||
| 3 | I | quercetin | OH | OH | OH | OH | OH | 6.10 | ||||
| 4 | I | baicalein | OH | OH | OH | 6.75 | ||||||
| 5 | I | luteolin | OH | OH | OH | OH | 27.22 | |||||
| 6 | III | isoliquiritigenin | OH | OH | OH | 36.29 | ||||||
| 7 | I | chrysin | OH | OH | 70.07 | |||||||
| 8 | II | dihydromyricetin | OH | OH | OH | OH | OH | OH | 85.95 | |||
| 9 | III | 4-hydroxychalcone | OH | 123.04 | ||||||||
| 10 | III | 4′-hydroxychalcone | OH | 199.10 | ||||||||
| 11 | II | hesperetin | OH | OH | OH | OCH3 | 286.60 | |||||
| 12 | II | naringin | OH | O-Rut | OH | >500 | ||||||
| 13 | II | hesperidin | OH | O-Rut | OH | OCH3 | >500 | |||||
| 14 | I | baicalin | OH | OH | O-Glu | >500 | ||||||
| 15 | I | rutin | O-Rut | OH | OH | OH | OH | >500 | ||||
| Flavonoids | T (K) | 1012 Kq (L·mol−1·s−1) | 104 Ka (L·mol−1) | n | ΔG (KJ·mol−1) | ΔH (KJ·mol−1) | ΔS (J·mol−1·K−1) |
|---|---|---|---|---|---|---|---|
| Myricetin | 298 | 9.6733 | 7.7786 | 0.9843 | −27.91 | −156.15 | −430.33 |
| 303 | 5.9984 | 2.7428 | 0.9386 | −25.76 | |||
| Dihydromyricetin | 298 | 3.1750 | 5.7637 | 1.0572 | −27.16 | −115.61 | −296.81 |
| 303 | 3.4092 | 2.6546 | 0.9764 | −25.68 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, C.; Liu, X.; Jiang, Z.; Geng, S.; Ma, H.; Liu, B. Interaction Mechanism of Flavonoids and α-Glucosidase: Experimental and Molecular Modelling Studies. Foods 2019, 8, 355. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8090355
He C, Liu X, Jiang Z, Geng S, Ma H, Liu B. Interaction Mechanism of Flavonoids and α-Glucosidase: Experimental and Molecular Modelling Studies. Foods. 2019; 8(9):355. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8090355
Chicago/Turabian StyleHe, Chengyun, Xiaoling Liu, Zhaojing Jiang, Sheng Geng, Hanjun Ma, and Benguo Liu. 2019. "Interaction Mechanism of Flavonoids and α-Glucosidase: Experimental and Molecular Modelling Studies" Foods 8, no. 9: 355. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8090355