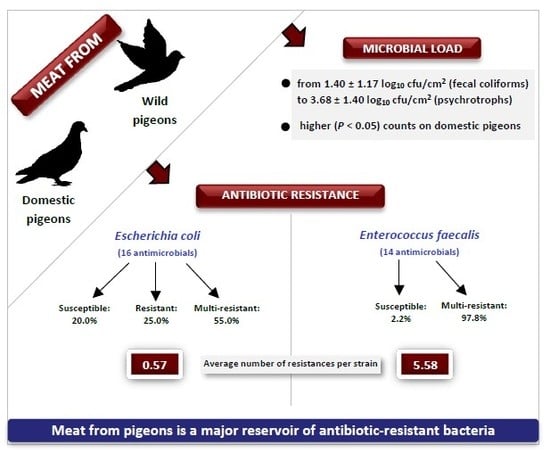
Microbial Load and Antibiotic Resistance Patterns of Escherichia coli and Enterococcus faecalis Isolates from the Meat of Wild and Domestic Pigeons
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Microbiological Determinations
2.3. Isolation and Identification of Escherichia coli and Enterococcus spp. Strains
2.4. Antibiotic Susceptibility Determination
2.5. Statistical Analysis
3. Results
3.1. Microbial Loads
3.2. Antibiotic Susceptibility in E. coli Strains
3.3. Antibiotic Susceptibility in E. faecalis Strains
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Game Meat—Production and Trade in the UNECE Region, FAO, Geneva. Available online: https://www.unece.org/fileadmin/DAM/timber/meetings/2018/20180321/game-meat-draft-2018-03.pdf (accessed on 14 September 2019).
- El Sector de la Avicultura de Carne en Cifras. Principales Indicadores Económicos. 2018. Ministerio de Agricultura, Pesca y Alimentación. Available online: https://www.mapa.gob.es/es/ganaderia/temas/produccion-y-mercados-ganaderos/indicadoresaviculturacarneparapublicar2018_tcm30-419674.pdf (accessed on 14 September 2019).
- European Commision. Commission regulation (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Off. J. Eur. Union 2005, L 338, 1–26. [Google Scholar]
- Del Río, E.; Panizo-Morán, M.; Prieto, M.; Alonso-Calleja, C.; Capita, R. Effect of various chemical decontamination treatments on natural microflora and sensory characteristics of poultry. Int. J. Food Microbiol. 2007, 115, 268–280. [Google Scholar] [CrossRef]
- Buzón-Durán, L.; Capita, R.; Alonso-Calleja, C. Microbial loads and antibiotic resistance patterns of Staphylococcus aureus in different types of raw poultry-based meat preparations. Poult. Sci. 2017, 96, 4046–4052. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Brzuszkiewicz, E.; Thurmer, A.; Schuldes, J.; Leimbach, A.; Liesegang, H.; Meyer, F.D.; Boelter, J.; Petersen, H.; Gottschalk, G.; Daniel, R. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. 2011, 193, 883–891. [Google Scholar] [CrossRef] [PubMed]
- EFSA; ECDC. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500, 262. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Cordero, J.; Molina-González, D.; Poeta, P.; Igrejas, G.; Alonso-Calleja, C.; Capita, R. Antimicrobial resistance and virulence genes in enterococci from wild game meat in Spain. Food Microbiol. 2016, 53, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance. Tackling the Burden in the European Union. Organization for Economic Coordination and Development (OECD). Available online: https://www.oecd.org/health/health-systems/AMR-Tackling-the-Burden-in-the-EU-OECD-ECDC-Briefing-Note-2019.pdf (accessed on 14 September 2019).
- EFSA; ECDC. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, e05598, 278. [Google Scholar] [CrossRef]
- Allen, S.E.; Boerlin, P.; Janecko, N.; Lumsden, J.S.; Barker, I.K.; Pearl, D.L.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario, Canada. Appl. Environ. Microbiol. 2011, 77, 882–888. [Google Scholar] [CrossRef]
- Santos, T.; Silva, N.; Igrejas, G.; Rodrigues, P.; Micael, J.; Rodrigues, T.; Resendes, R.; Goncalves, A.; Marinho, C.; Goncalves, D.; et al. Dissemination of antibiotic resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe 2013, 24, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Jay, J.M. A review of aerobic and psychrotrophic plate count procedures for fresh meat and poultry products. J. Food Prot. 2002, 65, 1200–1206. [Google Scholar] [CrossRef] [PubMed]
- Cousin, M.A.; Jay, J.M.; Vasavada, P.C. Psychrotrophic microorganisms. In Compendium of Methods for the Microbiological Examination of Foods; Downes, F.P., Ito, K., Eds.; American Public Health Association: Washington, DC, USA, 2001; pp. 159–166. [Google Scholar]
- Baird, R.M.; Corry, J.E.J.; Curtis, G.D.W. Pharmacopeia of culture media for food microbiology. Int. J. Food Microbiol. 1987, 5, 221–222. [Google Scholar]
- Anonymous. The Oxoid Manual; Oxoid: Basingstoke, UK, 1990. [Google Scholar]
- CLSI. M100 Performance Standars for Antimicrobial Susceptibility Testing, 29th ed.; Clinical and Laboratory Standards Institute: Pennsylvania, PA, USA, 2019. [Google Scholar]
- Capita, R.; Alonso-Calleja, C.; García-Fernández, M.C.; Moreno, B. Microbiological quality of retail poultry carcasses in Spain. J. Food Prot. 2001, 64, 1961–1966. [Google Scholar] [CrossRef]
- Álvarez-Astorga, M.; Capita, R.; Alonso-Calleja, C.; Moreno, B.; García-Fernández, M.C. Microbiological quality of retail chicken by-products in Spain. Meat Sci. 2002, 62, 45–50. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C.; García-Arias, M.T.; Moreno, B.; García-Fernández, M.C. Methods to detect the occurrence of various indicator bacteria on the surface of retail poultry in Spain. J. Food Sci. 2002, 67, 765–771. [Google Scholar] [CrossRef]
- Paulsen, P.; Smulders, F.J.M.; Hilbert, F. Salmonella in meat from hunted game: A central European perspective. Food Res. Int. 2008, 45, 609–616. [Google Scholar] [CrossRef]
- El-Ghareeb, W.R.; Morshdy, A.M.A.; Winkelmayer, R.; Paulsen, P. Microbiological condition and shelf life of meat from hunted game birds. Eur. J. Wildl. Res. 2009, 55, 317–323. [Google Scholar] [CrossRef]
- El-Dengawy, R.A.; Nassar, A.M. Investigation on the nutritive value and microbiological quality of wild quail carcasses. Nahrung 2001, 45, 50–54. [Google Scholar] [CrossRef]
- Mousa, M.M.; Salem, N.I.E.; Khalifa, A.M.A.; Abd-El-Hady, H.A.M.; El Gamel, A.M. Quality assessment of frozen quail in Kafr El Sheikl Governorate. Alex. J. Vet. Sci. 2016, 48, 57–61. [Google Scholar]
- Gill, C.O. Microbiological conditions of meats from large game animals and birds. Meat Sci. 2007, 77, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Costa, H.; Mafra, I.; Oliveira, M.B.P.P.; Amaral, J.S. Game: Types and composition. In The Encyclopedia of Food and Health; Caballero, B., Finglas, P., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; Volume 3, pp. 177–183. [Google Scholar]
- Patria, C.A.; Afnan, R.; Arief, I.I. Physical and microbiological qualities of kampong-broiler crossbred chickens meat raised in different stocking densities. Media Peternakan 2016, 39, 141–147. [Google Scholar] [CrossRef]
- Logue, C.M.; Sherwood, J.S.; Olah, P.A.; Elijah, L.M.; Dockter, M.R. The incidence of antimicrobial-resistant Salmonella spp. on freshly processed poultry from US Midwestern processing plants. J. Appl. Microbiol. 2003, 94, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, S.E. The relationship between antimicrobial resistance and patient outcomes: Mortality, length of hospital stay, and health care costs. Clin. Infect. Dis. 2006, 42 (Suppl. 2), S82–S89. [Google Scholar] [CrossRef]
- Tumbarello, M.; Spanu, T.; Di Bidino, R.; Marchetti, M.; Ruggeri, M.; Trecarichi, E.M.; De Pascale, G.; Proli, E.M.; Cauda, R.; Cicchetti, A.; et al. Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum-beta-lactamase production and inadequate initial antibiotic therapy. Antimicrobiol. Agents Chemother. 2010, 54, 4085–4091. [Google Scholar] [CrossRef]
- Guerra, B.; Junker, E.; Schroeter, A.; Malorny, B.; Lehmann, S.; Helmuth, R. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J. Antimicrob. Chemother. 2003, 52, 489–492. [Google Scholar] [CrossRef]
- Koo, H.; Woo, G. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 2011, 145, 407–413. [Google Scholar] [CrossRef]
- Álvarez-Fernández, E.; Cancelo, A.; Díaz-Vega, C.; Capita, R.; Alonso-Calleja, C. Antimicrobial resistance in E. coli isolates from conventionally and organically reared poultry: A comparison of agar disc diffusion and Sensi Test Gram-negative methods. Food Control 2013, 30, 227–234. [Google Scholar] [CrossRef]
- Capita, R.; Álvarez-Fernández, E.; Fernández-Buelta, E.; Manteca, J.; Alonso-Calleja, C. Decontamination treatments can increase the prevalence of resistance to antibiotics of Escherichia coli naturally present on poultry. Food Microbiol. 2013, 34, 112–117. [Google Scholar] [CrossRef]
- Ojer-Usoz, E.; González, D.; Vitas, A.I.; Leiva, J.; García-Jalón, I.; Febles-Casquero, A.; Escolano, M. Prevalence of extended-spectrum-beta-lactamase producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013, 93, 316–321. [Google Scholar] [CrossRef]
- Hussain, A.; Shaik, S.; Ranjan, A.; Nandanwar, N.; Tiwari, S.K.; Majid, M.; Baddam, R.; Qureshi, I.A.; Semmler, T.; Wieler, L.H.; et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. Microbiol. 2017, 8, 2120. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Grande, H.; Weaver, B.; Papp, K.; Horwinski, J.; Koch, B.; Hungate, B.A.; Liu, C.M.; et al. Antibiotic-resistant Escherichia coli from retail poultry meat with different antibiotic use claims. BMC Microbiol. 2018, 18, 174. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, N.; Oka, C.; Sato, G. Isolation of citrate-positive variants of Escherichia coli from domestic pigeons, pigs, cattle, and horses. Appl. Environ. Microbiol. 1978, 36, 217–222. [Google Scholar] [PubMed]
- Kimpe, A.; Decostere, A.; Martel, A.; Haesebrouck, F.; Devriese, L.A. Prevalence of antimicrobial resistance among pigeon isolates of Streptococcus gallolyticus, Escherichia coli and Salmonella enterica serotype Typhimurium. Avian Pathol. 2002, 31, 393–397. [Google Scholar] [CrossRef]
- Costa, D.; Poeta, P.; Saenz, Y.; Vinue, L.; Rojo-Bezares, B.; Jouini, A.; Zarazaga, M.; Rodrigues, M.J.; Torres, C. Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 2006, 58, 1311–1312. [Google Scholar] [CrossRef]
- Radhouani, H.; Poeta, P.; Igrejas, G.; Goncalves, A.; Vinue, L.; Torres, C. Antimicrobial resistance and phylogenetic groups in isolates of Escherichia coli from seagulls at the Berlengas nature reserve. Vet. Rec. 2009, 165, 138–142. [Google Scholar] [CrossRef]
- Silva, V.L.; Nicoli, J.R.; Nascimento, T.C.; Diniz, C.G. Diarrheagenic Escherichia coli strains recovered from urban pigeons (Columba livia) in Brazil and their antimicrobial susceptibility patterns. Curr. Microbiol. 2009, 59, 302–308. [Google Scholar] [CrossRef]
- Radimersky, T.; Frolkova, P.; Janoszowska, D.; Dolejska, M.; Svec, P.; Roubalova, E.; Cikova, P.; Cizek, A.; Literak, I. Antibiotic resistance in faecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. J. Appl. Microbiol. 2010, 109, 1687–1695. [Google Scholar] [CrossRef]
- Radhouani, H.; Igrejas, G.; Gonçalves, A.; Pacheco, R.; Monteiro, R.; Sargo, R.; Brito, F.; Torres, C.; Poeta, P. Antimicrobial resistance and virulence genes in Escherichia coli and enterococci from red foxes (Vulpes vulpes). Anaerobe 2013, 23, 82–86. [Google Scholar] [CrossRef]
- Hasan, B.; Islam, K.; Ahsan, M.; Hossain, Z.; Rashid, M.; Talukder, B.; Ahmed, K.U.; Olsen, B.; Abul Kashem, M. Fecal carriage of multi-drug resistant and extended spectrum beta-lactamases producing E. coli in household pigeons, Bangladesh. Vet. Microbiol. 2014, 168, 221–224. [Google Scholar] [CrossRef]
- Stenzel, T.; Bancerz-Kisiel, A.; Tykałowski, B.; Śmiałek, M.; Pestka, D.; Koncicki, A. Antimicrobial resistance in bacteria isolated from pigeons in Poland. Pol. J. Vet. Sci. 2014, 17, 169–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chindamba, L.; Korsten, L. Antibiotic resistance in Escherichia coli isolates from roof-harvested rainwater tanks and urban pigeon faeces as the likely source of contamination. Environ. Monit. Assess. 2015, 187, 405. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Peña, K.; Esperón, F.; Torres-Mejía, A.M.; de la Torre, A.; de la Cruz, E.; Jimenez-Soto, M. Antimicrobial resistance genes in pigeons from public Parks in Costa Rica. Zoonoses Public Health 2017, 65, e23–e30. [Google Scholar] [CrossRef] [PubMed]
- Zigo, F.; Takac, L.; Zigova, M.; Takacova, J.; Vasi, M. Occurrence of antibiotic-resistant bacterial strains isolated in carrier pigeons during the race season. J. Chem. Pharm. Sci. 2017, 10, 10–13. [Google Scholar]
- Hata, A.; Shibahara, T.; Yamamoto, H.; Fujitani, N. Antimicrobial resistance of Enterobacteriaceae in feral pigeons living in the Kanto region of Japan. Int. J. Anal. Bio-Sci. 2018, 6, 10–18. [Google Scholar]
- Ngozi, A.; Agabus, N.; Eucharia, O.; Onyinyechi, U.-I.; Abraham, E.; Chika, E.; Ifeanyichukwu, I. A three-year study on the prevalence and antibiotic susceptibility pattern of Escherichia coli isolated from cloacal swabs of wild and domestic birds in Ebonyi State, Nigeria. EC Microbiol. 2018, 14, 266–273. [Google Scholar]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019; Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf?ua=1 (accessed on 14 October 2019).
- World Organization for Animal Health. OIE List of Antimicrobial Agents of Veterinary Importance; World Organization for Animal Health: Paris, France, 2019; Available online: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/A_OIE_List_antimicrobials_July2019.pdf (accessed on 14 October 2019).
- Navarro-González, N.; Casas-Díaz, E.; Porrero, C.M.; Mateos, A.; Domínguez, L.; Lavín, S.; Serrano, E. Food-borne zoonotic pathogens and antimicrobial resistance of indicator bacteria in urban wild boars in Barcelona, Spain. Vet. Microbiol. 2013, 167, 686–689. [Google Scholar] [CrossRef]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Gomes, B.C.; Esteves, C.T.; Palazzo, I.C.V.; Darini, A.L.C.; Felis, G.E.; Sechi, L.A.; Franco, B.D.G.M.; De Martinis, E.C.P. Prevalence and characterization of Enterococcus spp. isolated from Brazilian foods. Food Microbiol. 2008, 25, 668–675. [Google Scholar] [CrossRef]
- Pesavento, G.; Calonico, C.; Ducci, B.; Magnanini, A.; Lo Nostro, A. Prevalence and antibiotic resistance of Enterococcus spp. isolated from retail cheese, ready-to-eat salads, ham, and raw meat. Food Microbiol. 2014, 41, 1–7. [Google Scholar] [CrossRef]
- Valenzuela, A.; Omar, N.B.; Abriouel, H.; López, R.L.; Ortega, E.; Cañamero, M.M.; Gálvez, A. Risk factors. Risk factors in enterococci isolated from foods in Morocco; determination of antimicrobial resistance and incidence of virulent traits. Food Chem. Toxicol. 2008, 46, 2648–2652. [Google Scholar] [CrossRef] [PubMed]
- Poeta, P.; Costa, D.; Igrejas, G.; Rodrigues, J.; Torres, C. Phenotypic and genotypic characterization of antimicrobial resistance in faecal enterococci from wild boars (Sus scrofa). Vet. Microbiol. 2007, 125, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.; Igrejas, G.; Figueiredo, N.; Gonçalves, A.; Radhouani, H.; Rodrigues, J. Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 2010, 408, 4871–4876. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Ramos, E.; Molina-González, D.; Blanco-Morán, S.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Prevalence, antimicrobial resistance, and genotypic characterization of vancomycin-resistant enterococci in meat preparations. J. Food Prot. 2016, 79, 748–756. [Google Scholar] [CrossRef] [PubMed]
- De Briyne, N.; Atkinson, J.; Pokludová, L.; Borriello, S.P. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014, 175, 325. [Google Scholar] [CrossRef]
- Cameron, A.; McAllister, A.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Doming, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef]




| Microbial Group | Culture Medium | Incubation | Reference | |
|---|---|---|---|---|
| T (°C) | Time | |||
| Aerobic plate count (APC) 1 | Plate count agar (PCA) | 30 | 72 h | [15] |
| Psychrotrophs 1 | Plate count agar (PCA) | 7 | 10 d | [16] |
| Enterobacteriaceae2,3 | Violet red bile glucose agar (VRBGA) | 35 | 24 h | [17] |
| Fecal coliforms 2,3 | Violet red bile agar (VRBA) | 44 | 24 h | [17] |
| Enterococci 2 | Kanamycin aesculin azide (KEA) agar | 42 | 24 h | [17] |
| Micrococcaceae1 | Mannitol salt agar (MSA) | 35 | 24–48 h | [18] |
| Lactic acid bacteria 2 | De Man, Rogosa and Sharpe (MRS) agar | 30 | 72 h | [17] |
| Brochothrix thermosphacta1 | Streptomycin thallous acetate actidione (STAA) agar | 25 | 48 h | [18] |
| Type of Pigeon | |||
|---|---|---|---|
| Microbial Group | Total | Domestic | Wild |
| Aerobic plate count (APC) | 3.16 ± 0.95 c | 3.76 ± 0.92 bb | 2.90 ± 0.84 acd |
| Psychrotrophs | 3.68 ± 1.40 d | 3.84 ± 2.04 ab | 3.61 ± 1.04 ae |
| Enterobacteriaceae | 1.56 ± 1.25 a | 2.06 ± 1.19 ba | 1.35 ± 1.23 aab |
| Fecal coliforms | 1.40 ± 1.17 a | 2.11 ± 1.02 ba | 1.10 ± 1.11 aa |
| Enterococci | 1.82 ± 1.12 a | 2.17 ± 0.85 aa | 1.67 ± 1.20 ab |
| Micrococcaceae | 2.73 ± 1.16 b | 3.12 ± 1.05 bb | 2.56 ± 1.17 ac |
| Lactic acid bacteria (LAB) | 3.40 ± 1.04 cd | 3.81 ± 0.93 bb | 3.23 ± 1.05 ade |
| Brochothrix thermosphacta | 1.53 ± 2.08 a | 1.47 ± 2.17 aa | 1.56 ± 2.05 aab |
| Antibiotic | Strains from Domestic Pigeons (n = 32) | Strains from Wild Pigeons (n = 28) | All Strains (n = 60) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Isolates | % of Resistant Strains | % of Resistant and Intermediate Strains | Number of Isolates | % of Resistant Strains | % of Resistant and Intermediate Strains | Number of Isolates | % of Resistant Strains | % of Resistant and Intermediate Strains | |||||||
| S | I | R | S | I | R | S | I | R | |||||||
| AMC | 28 | 4 | 0 | 0.00% | 12.50% | 20 | 6 | 2 | 7.14% | 28.57% | 48 | 10 | 2 | 3.33 | 20.00% |
| AMP | 25 | 7 | 0 | 0.00% | 21.88% | 19 | 5 | 4 | 14.29% | 32.14% | 44 | 12 | 4 | 6.67 | 26.67% |
| CAZ | 31 | 1 | 0 | 0.00% | 3.13% | 26 | 2 | 0 | 0.00% | 7.14% | 57 | 3 | 0 | 0.00 | 5.00% |
| CTX | 27 | 4 | 1 | 3.13% | 15.63% | 25 | 3 | 0 | 0.00% | 10.71% | 52 | 7 | 1 | 1.67 | 13.33% |
| FOX | 32 | 0 | 0 | 0.00% | 0.00% | 24 | 1 | 3 | 10.71% | 14.29% | 56 | 1 | 3 | 5.00 | 6.67% |
| IPM | 32 | 0 | 0 | 0.00% | 0.00% | 28 | 0 | 0 | 0.00% | 0.00% | 60 | 0 | 0 | 0.00 | 0.00% |
| ATM | 28 | 3 | 1 | 3.13% | 12.50% | 23 | 1 | 4 | 14.29% | 17.86% | 51 | 4 | 5 | 8.33 | 15.00% |
| NA | 32 | 0 | 0 | 0.00% | 0.00% | 25 | 2 | 1 | 3.57% | 10.71% | 57 | 2 | 1 | 1.67 | 5.00% |
| CIP | 32 | 0 | 0 | 0.00% | 0.00% | 27 | 1 | 0 | 0.00% | 3.57% | 59 | 1 | 0 | 0.00 | 1.67% |
| C | 30 | 2 | 0 | 0.00% | 6.25% | 27 | 1 | 0 | 0.00% | 3.57% | 57 | 3 | 0 | 0.00 | 5.00% |
| CN | 30 | 2 | 0 | 0.00% | 6.25% | 23 | 0 | 5 | 17.86% | 17.86% | 53 | 2 | 5 | 8.33 | 11.67% |
| AK | 31 | 1 | 0 | 0.00% | 3.13% | 22 | 2 | 4 | 14.29% | 21.43% | 53 | 3 | 4 | 6.67 | 11.67% |
| STR | 15 | 15 | 2 | 6.25% | 53.13% | 12 | 15 | 1 | 3.57% | 57.14% | 27 | 30 | 3 | 5.00 | 55.00% |
| TOB | 27 | 4 | 1 | 3.13% | 15.63% | 21 | 4 | 3 | 10.71% | 25.00% | 48 | 8 | 4 | 6.67 | 20.00% |
| TE | 31 | 0 | 1 | 3.13% | 3.13% | 25 | 2 | 1 | 3.57% | 10.71% | 56 | 2 | 2 | 3.33 | 6.67% |
| SXT | 32 | 0 | 0 | 0.00% | 0.00% | 28 | 0 | 0 | 0.00% | 0.00% | 60 | 0 | 0 | 0.00 | 0.00% |
| Antibiotic | Strains from Domestic Pigeons (n = 16) | Strains from Wild Pigeons (n = 29) | All Strains (n = 45) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Isolates | % of Resistant Strains | % of Resistant and Intermediate Strains | Number of Isolates | % of Resistant Strains | % of Resistant and Intermediate Strains | Number of Isolates | % of Resistant Strains | % of Resistant and Intermediate Strains | |||||||
| S | I | R | S | I | R | S | I | R | |||||||
| AMP | 4 | 10 | 2 | 12,50% | 75.00% | 13 | 8 | 8 | 27.59% | 55.17% | 17 | 18 | 10 | 22.22% | 62.22% |
| CIP | 0 | 4 | 12 | 75.00% | 100% | 3 | 13 | 13 | 44.83% | 89.66% | 3 | 17 | 25 | 55.56% | 93.33% |
| C | 7 | 9 | 0 | 0.00% | 56.25% | 3 | 25 | 1 | 3.45% | 89.66% | 10 | 34 | 1 | 2.22% | 77.78% |
| CN | 3 | 6 | 7 | 43.75% | 81.25% | 3 | 3 | 23 | 79.31% | 89.66% | 6 | 9 | 30 | 66.67% | 86.67% |
| K | 0 | 2 | 14 | 87.50% | 100% | 7 | 4 | 18 | 62.07% | 75.86% | 7 | 6 | 32 | 71.11% | 84.44% |
| STR | 0 | 1 | 15 | 93.75% | 100% | 3 | 1 | 25 | 86.21% | 89.66% | 3 | 2 | 40 | 88.89% | 93.33% |
| E | 0 | 9 | 7 | 43.75% | 100% | 1 | 19 | 9 | 31.03% | 96.55% | 1 | 28 | 16 | 35.56% | 97.78% |
| FOS | 7 | 6 | 3 | 18.75% | 56.25% | 14 | 11 | 4 | 13.79% | 51.72% | 21 | 17 | 7 | 15.56% | 53.33% |
| F | 3 | 5 | 8 | 50.00% | 81.25% | 4 | 5 | 20 | 68.97% | 86.21% | 7 | 10 | 28 | 62.22% | 84.44% |
| RD | 7 | 3 | 6 | 37.50% | 56.25% | 3 | 6 | 20 | 68.97% | 89.66% | 10 | 9 | 26 | 57.78% | 77.78% |
| TEC | 7 | 9 | 0 | 0.00% | 56.25% | 22 | 6 | 1 | 3.45% | 24.14% | 29 | 15 | 1 | 2.22% | 35.56% |
| VA | 6 | 9 | 1 | 6.25% | 62.50% | 5 | 19 | 5 | 17.24% | 82.76% | 11 | 28 | 6 | 13.33% | 75.56% |
| TE | 0 | 5 | 11 | 68.75% | 100% | 2 | 10 | 17 | 58.62% | 93.10% | 2 | 15 | 28 | 62.22% | 95.56% |
| SXT | 11 | 5 | 0 | 0.00% | 31.25% | 20 | 8 | 1 | 3.45% | 31.03% | 31 | 13 | 1 | 2.22% | 31.11% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordero, J.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Microbial Load and Antibiotic Resistance Patterns of Escherichia coli and Enterococcus faecalis Isolates from the Meat of Wild and Domestic Pigeons. Foods 2019, 8, 536. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8110536
Cordero J, Alonso-Calleja C, García-Fernández C, Capita R. Microbial Load and Antibiotic Resistance Patterns of Escherichia coli and Enterococcus faecalis Isolates from the Meat of Wild and Domestic Pigeons. Foods. 2019; 8(11):536. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8110536
Chicago/Turabian StyleCordero, Jorge, Carlos Alonso-Calleja, Camino García-Fernández, and Rosa Capita. 2019. "Microbial Load and Antibiotic Resistance Patterns of Escherichia coli and Enterococcus faecalis Isolates from the Meat of Wild and Domestic Pigeons" Foods 8, no. 11: 536. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8110536