Bioprinted Three-Dimensional Cell-Laden Hydrogels to Evaluate Adipocyte-Breast Cancer Cell Interactions
Abstract
:1. Introduction
2. Results and Discussion
2.1. Hydrogel Bioink Screening
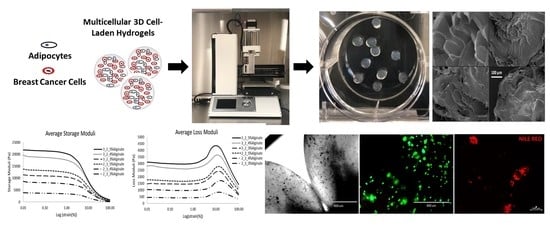
2.2. Characterization of Bioprinted 3D Hydrogels
2.2.1. Rheological Characterization
2.2.2. Bioprinted 3D Hydrogel Degradation and Morphology
2.3. Preliminary Screening of Culture Medium
2.3.1. Two-Dimensional Cell Viability and Metabolic Activity
2.3.2. Two-Dimensional Adipogenic Potential
2.4. Evaluation of 3D Bioprinted Cell-Laden Hydrogels
2.4.1. Assessment of 3D Cell-Laden Hydrogel Monocultures
2.4.2. Assessment of 3D Cell-Laden Hydrogel Co-Cultures
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Bioprinter Optimization
4.3. Hydrogel Bioink Screening
4.3.1. Three-Dimensional Hydrogel Fabrication and Characterization
4.3.2. Rheological Characterization
4.3.3. Three-Dimensional Hydrogel Degradation
4.3.4. Scanning Electron Microscopy
4.4. Cell Expansion and Seeding
4.5. Preliminary Screening of Culture Medium
4.6. Bioprinting of 3D Cell-laden Hydrogels
4.7. In Vitro Cell Viability and Metabolic Activity Assessment
4.8. Adipogenic Potential
4.9. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeSantis, C.; Ma, J.; Bryan, L.; Jemal, A. Breast cancer statistics, 2013. CA Cancer J. Clin. 2014, 64, 52–62. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, C.; Siegel, R.; Bandi, P.; Jemal, A. Breast cancer statistics, 2011. CA Cancer J. Clin. 2011, 61, 408–418. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorincz, A.; Sukumar, S. Molecular links between obesity and breast cancer. Endocr. Relat. Cancer 2006, 13, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Cleary, M.P.; Grossmann, M.E. Obesity and breast cancer: the estrogen connection. Endocrinology 2009, 150, 2537–2542. [Google Scholar] [CrossRef]
- Vandeweyer, E.; Hertens, D. Quantification of glands and fat in breast tissue: an experimental determination. Ann. Anat. 2002, 184, 181–184. [Google Scholar] [CrossRef]
- Kim, J.B.; Stein, R.; O’hare, M.J. Three-dimensional in vitro tissue culture models of breast cancer—A review. Breast Cancer Res. Treat. 2004, 85, 281–291. [Google Scholar] [CrossRef]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef] [Green Version]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T. Comparison of 2D-and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.M.; Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bougaret, L.; Delort, L.; Billard, H.; Le Huede, C.; Boby, C.; De la Foye, A.; Rossary, A.; Mojallal, A.; Damour, O.; Auxenfans, C.; et al. Adipocyte/breast cancer cell crosstalk in obesity interferes with the anti-proliferative efficacy of tamoxifen. PloS ONE 2018, 13, e0191571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, G.K.; Pirie, K.; Beral, V.; Green, J.; Spencer, E.; Bull, D. Cancer incidence and mortality in relation to body mass index in the Million Women Study: cohort study. BMJ 2007, 335, 1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celis, J.E.; Moreira, J.M.; Cabezón, T.; Gromov, P.; Friis, E.; Rank, F.; Gromova, I. Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients: toward dissecting the molecular circuitry of epithelial-adipocyte stromal cell interactions. Mol. Cell. Proteom. 2005, 4, 492–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perera, C.N.; Spalding, H.S.; Mohammed, S.I.; Camarillo, I.G. Identification of proteins secreted from leptin stimulated MCF-7 breast cancer cells: a dual proteomic approach. Exp. Biol. Med. (Maywood) 2008, 233, 708–720. [Google Scholar] [CrossRef]
- Duong, M.N.; Geneste, A.; Fallone, F.; Li, X.; Dumontet, C.; Muller, C. The fat and the bad: Mature adipocytes, key actors in tumor progression and resistance. Oncotarget 2017, 8, 57622. [Google Scholar] [CrossRef] [Green Version]
- Bochet, L.; Escourrou, G.; Valet, P.; Muller, C. Unraveling the obesity and breast cancer links: A role for cancer-associated adipocytes? In Adipose Tissue Development; Karger Publishers: Basel, Switzerland, 2010; Volume 19, pp. 45–52. [Google Scholar]
- Dirat, B.; Bochet, L.; Dabek, M.; Daviaud, D.; Dauvillier, S.; Majed, B.; Wang, Y.Y.; Meulle, A.; Salles, B.; Le Gonidec, S. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011, 71, 2455–2465. [Google Scholar] [CrossRef] [Green Version]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773. [Google Scholar] [CrossRef]
- Arslan-Yildiz, A.; El Assal, R.; Chen, P.; Guven, S.; Inci, F.; Demirci, U. Towards artificial tissue models: past, present, and future of 3D bioprinting. Biofabrication 2016, 8, 014103. [Google Scholar] [CrossRef]
- Asghar, W.; El Assal, R.; Shafiee, H.; Pitteri, S.; Paulmurugan, R.; Demirci, U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today (Kidlington) 2015, 18, 539–553. [Google Scholar] [CrossRef]
- Ozbolat, I.T.; Peng, W.; Ozbolat, V. Application areas of 3D bioprinting. Drug Discov. Today 2016, 21, 1257–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pati, F.; Gantelius, J.; Svahn, H.A. 3D Bioprinting of Tissue/Organ Models. Angew. Chem. Int. Ed. Engl. 2016, 55, 4650–4665. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, W.; Nowicki, M.; Miao, S.; Cui, H.; Holmes, B.; Glazer, R.I.; Zhang, L.G. 3D Bioprinting a Cell-Laden Bone Matrix for Breast Cancer Metastasis Study. ACS Appl. Mater. Interfaces 2016, 8, 30017–30026. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, S.; Onal, S.; Yu, C.H.; Zhao, J.J.; Tasoglu, S. Bioprinting for cancer research. Trends Biotechnol. 2015, 33, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Pusch, K.; Hinton, T.J.; Feinberg, A.W. Large volume syringe pump extruder for desktop 3D printers. HardwareX 2018, 3, 49–61. [Google Scholar] [CrossRef]
- Stanton, M.; Samitier, J.; Sanchez, S. Bioprinting of 3D hydrogels. Lab. Chip 2015, 15, 3111–3115. [Google Scholar] [CrossRef] [Green Version]
- Gomillion, C.T.; Yang, C.-C.; Dréau, D.; Burg, K.J. Engineered Composites for 3D Mammary Tissue Systems; Taylor and Francis: Abingdon, UK, 2017. [Google Scholar]
- Krouskop, T.A.; Wheeler, T.M.; Kallel, F.; Garra, B.S.; Hall, T. Elastic moduli of breast and prostate tissues under compression. Ultrason. Imaging 1998, 20, 260–274. [Google Scholar] [CrossRef]
- McKnight, A.L.; Kugel, J.L.; Rossman, P.J.; Manduca, A.; Hartmann, L.C.; Ehman, R.L. MR elastography of breast cancer: preliminary results. AJR Am. J. Roentgenol. 2002, 178, 1411–1417. [Google Scholar] [CrossRef]
- Sarvazyan, A.P.; Rudenko, O.V.; Swanson, S.D.; Fowlkes, J.B.; Emelianov, S.Y. Shear wave elasticity imaging: a new ultrasonic technology of medical diagnostics. Ultrasound Med. Biol. 1998, 24, 1419–1435. [Google Scholar] [CrossRef]
- Chang, J.M.; Park, I.A.; Lee, S.H.; Kim, W.H.; Bae, M.S.; Koo, H.R.; Yi, A.; Kim, S.J.; Cho, N.; Moon, W.K. Stiffness of tumours measured by shear-wave elastography correlated with subtypes of breast cancer. Eur. Radiol. 2013, 23, 2450–2458. [Google Scholar] [CrossRef]
- Kong, H.J.; Wong, E.; Mooney, D.J. Independent Control of Rigidity and Toughness of Polymeric Hydrogels. Macromolecules 2003, 36, 4582–4588. [Google Scholar] [CrossRef]
- Shengmao, L.; Linxia, G. Influence of Crosslink Density and Stiffness on Mechanical Properties of Type I Collagen Gel. Materials 2015, 8, 551–560. [Google Scholar] [CrossRef]
- Kershaw, E.E.; Flier, J. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Galic, S.; Oakhill, J.S.; Steinberg, G.R. Adipose tissue as an endocrine organ. Mol. Cell. Endocrinol. 2010, 316, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.; Rodeheffer, M.S.; Rosen, C.J.; Horowitz, M.C. Adipose Tissue-Residing Progenitors (Adipocyte Lineage Progenitors and Adipose-Derived Stem Cells (ADSC)). Curr. Mol. Biol. Rep. 2015, 1, 101–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Zou, J.; Wang, W.; Nie, Y.; Tung, W.-T.; Ma, N.; Lendlein, A. Dedifferentiation of mature adipocytes with periodic exposure to cold. Clin. Hemorheol. Microcirc. 2019, 77, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Petan, T.; Jarc, E.; Jusović, M. Lipid droplets in cancer: guardians of fat in a stressful world. Molecules 2018, 23, 1941. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Celli, J.; Rizvi, I.; Moon, S.; Hasan, T.; Demirci, U. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol. J. 2011, 6, 204–212. [Google Scholar] [CrossRef]
- Hockel, M.; Vaupel, P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 2001, 93, 266–276. [Google Scholar] [CrossRef] [Green Version]








| Hydrogel Material | Solvent | Crosslinker | Structure Maintained (Y/N) | Significant Degradation Observed (Y/N) | Cells Viable After Crosslinking (Y/N) |
|---|---|---|---|---|---|
| Agarose | Deionized H2O | None/Cooled PBS | Y | Y | Y |
| Chitosan | 1% Glacial Acetic Acid | 0.5 N Sodium Hydroxide | Y | N | N |
| Pectin | Deionized H2O | 1 M Calcium Chloride | N | Y | Y |
| Alginate | Deionized H2O | 1 M Calcium Chloride | Y | Y | Y |
| Gelatin | Deionized H2O | None/Cooled PBS | Y | Y | Y |
| Alginate/Gelatin | Deionized H2O | 0.05 M Calcium Chloride | Y | N | Y |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaji, S.; Al-Saleh, J.; Gomillion, C.T. Bioprinted Three-Dimensional Cell-Laden Hydrogels to Evaluate Adipocyte-Breast Cancer Cell Interactions. Gels 2020, 6, 10. https://0-doi-org.brum.beds.ac.uk/10.3390/gels6010010
Chaji S, Al-Saleh J, Gomillion CT. Bioprinted Three-Dimensional Cell-Laden Hydrogels to Evaluate Adipocyte-Breast Cancer Cell Interactions. Gels. 2020; 6(1):10. https://0-doi-org.brum.beds.ac.uk/10.3390/gels6010010
Chicago/Turabian StyleChaji, Sarah, Jenna Al-Saleh, and Cheryl T. Gomillion. 2020. "Bioprinted Three-Dimensional Cell-Laden Hydrogels to Evaluate Adipocyte-Breast Cancer Cell Interactions" Gels 6, no. 1: 10. https://0-doi-org.brum.beds.ac.uk/10.3390/gels6010010