Exome Evaluation of Autism-Associated Genes in Amazon American Populations
Abstract
:1. Introduction
2. Materials and Methods
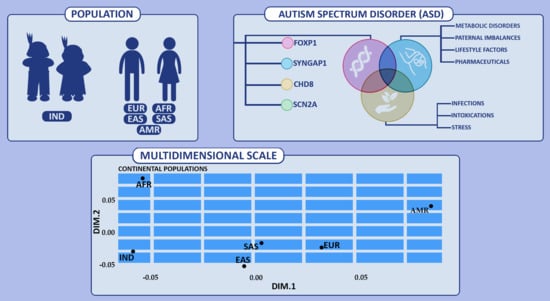
2.1. Study and Reference Populations
2.2. Extraction of the DNA and Preparation of the Exome Library
2.3. Bioinformatics Analysis
2.4. Statistical Analysis
2.5. Selection of Genes and Variants
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Transtorno Do Espectro Autista—OPAS/OMS | Organização Pan-Americana da Saúde. Available online: https://www.paho.org/pt/topicos/transtorno-do-espectro-autista (accessed on 1 September 2021).
- Elsabbagh, M.; Divan, G.; Koh, Y.-J.; Kim, Y.S.; Kauchali, S.; Marcín, C.; Montiel-Nava, C.; Patel, V.; Paula, C.S.; Wang, C.; et al. Global Prevalence of Autism and Other Pervasive Developmental Disorders. Autism Res. 2012, 5, 160–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, B.; Arciuli, J. Indigenous Australians with Autism: A Scoping Review. Autism 2020, 24, 1031–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron-Cohen, S. Leo Kanner, Hans Asperger, and the Discovery of Autism. Lancet 2015, 386, 1329–1330. [Google Scholar] [CrossRef] [Green Version]
- Kanner, L. Autistic Disturbances of Affective Contact. Nerv. Child 1943, 2, 217–250. [Google Scholar]
- Silberman, S.; Sacks, O. NeuroTribes the Legacy of Autism and How to Think Smarter about People Who Think Differently; Allen & Unwin: London, UK, 2015. [Google Scholar]
- Bai, D.; Yip, B.H.K.; Windham, G.C.; Sourander, A.; Francis, R.; Yoffe, R.; Glasson, E.; Mahjani, B.; Suominen, A.; Leonard, H.; et al. Association of Genetic and Environmental Factors with Autism in a 5-Country Cohort. JAMA Psychiatry 2019, 76, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.R.; Gonda, X.; Tarazi, F.I. Autism Spectrum Disorder: Classification, Diagnosis and Therapy. Pharmacol. Ther. 2018, 190, 91–104. [Google Scholar] [CrossRef]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.-Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef]
- The 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning. 1; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Rodrigues, J.C.G.; Fernandes, M.R.; Guerreiro, J.F.; da Silva, A.L.D.C.; Ribeiro-Dos-Santos, Â.; Santos, S.; Santos, N.P.C.D. Polymorphisms of ADME-Related Genes and Their Implications for Drug Safety and Efficacy in Amazonian Amerindians. Sci. Rep. 2019, 9, 7201. [Google Scholar] [CrossRef]
- Maenner, M.J. Prevalence of Autism Spectrum Disorder among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR. Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef]
- da Silva Montenegro, E.M.; Costa, C.S.; Campos, G.; Scliar, M.; de Almeida, T.F.; Zachi, E.C.; Silva, I.M.W.; Chan, A.J.S.; Zarrei, M.; Lourenço, N.C.V.; et al. Meta-Analyses Support Previous and Novel Autism Candidate Genes: Outcomes of an Unexplored Brazilian Cohort. Autism Res. Off. J. Int. Soc. Autism Res. 2020, 13, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Shochet, I.M.; Orr, J.A.; Kelly, R.L.; Wurfl, A.M.; Saggers, B.R.; Carrington, S.B. Psychosocial Resources Developed and Trialled for Indigenous People with Autism Spectrum Disorder and Their Caregivers: A Systematic Review and Catalogue. Int. J. Equity Health 2020, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Larsson, H.; Hultman, C.M.; Reichenberg, A. The Familial Risk of Autism. JAMA 2014, 311, 1770. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.P.C.; Ribeiro-Rodrigues, E.M.; Ribeiro-dos-Santos, Â.K.C.; Pereira, R.; Gusmão, L.; Amorim, A.; Guerreiro, J.F.; Zago, M.A.; Matte, C.; Hutz, M.H.; et al. Assessing Individual Interethnic Admixture and Population Substructure Using a 48-Insertion-Deletion (INSEL) Ancestry-Informative Marker (AIM) Panel. Hum. Mutat. 2010, 31, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues de Moura, R.; Coelho, A.V.C.; de Queiroz Balbino, V.; Crovella, S.; Brandão, L.A.C. Meta-Analysis of Brazilian Genetic Admixture and Comparison with Other Latin America Countries. Am. J. Hum. Biol. 2015, 27, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, M.; Ramzan, K. A de Novo Variant of CHD8 in a Patient with Autism Spectrum Disorder. Discoveries 2020, 8, e107. [Google Scholar] [CrossRef]
- Hoffmann, A.; Spengler, D. Chromatin Remodeler CHD8 in Autism and Brain Development. J. Clin. Med. 2021, 10, 366. [Google Scholar] [CrossRef]
- Tavassoli, T.; Kolevzon, A.; Wang, A.T.; Curchack-Lichtin, J.; Halpern, D.; Schwartz, L.; Soffes, S.; Bush, L.; Grodberg, D.; Cai, G.; et al. De Novo SCN2A Splice Site Mutation in a Boy with Autism Spectrum Disorder. BMC Med. Genet. 2014, 15, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Varghese, M.; Keshav, N.; Jacot-Descombes, S.; Warda, T.; Wicinski, B.; Dickstein, D.L.; Harony-Nicolas, H.; De Rubeis, S.; Drapeau, E.; Buxbaum, J.D.; et al. Autism Spectrum Disorder: Neuropathology and Animal Models. Acta Neuropathol. 2017, 134, 537–566. [Google Scholar] [CrossRef]
- Bowers, J.M.; Konopka, G. The Role of the FOXP Family of Transcription Factors in ASD. Dis. Mark. 2012, 33, 251–260. [Google Scholar] [CrossRef]
- Siper, P.M.; De Rubeis, S.; Trelles, M.d.P.; Durkin, A.; Di Marino, D.; Muratet, F.; Frank, Y.; Lozano, R.; Eichler, E.E.; Kelly, M.; et al. Prospective Investigation of FOXP1 Syndrome. Mol. Autism 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skoglund, P.; Reich, D. A Genomic View of the Peopling of the Americas. Curr. Opin. Genet. Dev. 2016, 41, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffecker, J.F.; Elias, S.A.; O’Rourke, D.H.; Scott, G.R.; Bigelow, N.H. Beringia and the Global Dispersal of Modern Humans. Evol. Anthropol. Issues News Rev. 2016, 25, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Reich, D.; Patterson, N.; Campbell, D.; Tandon, A.; Mazieres, S.; Ray, N.; Parra, M.V.; Rojas, W.; Duque, C.; Mesa, N.; et al. Reconstructing Native American Population History. Nature 2012, 488, 370–374. [Google Scholar] [CrossRef]
- Gravel, S.; Zakharia, F.; Moreno-Estrada, A.; Byrnes, J.K.; Muzzio, M.; Rodriguez-Flores, J.L.; Kenny, E.E.; Gignoux, C.R.; Maples, B.K.; Guiblet, W.; et al. Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data. PLoS Genet. 2013, 9, e1004023. [Google Scholar] [CrossRef]
- Rodrigues, J.C.G.; Souza, T.P.D.; Pastana, L.F.; Ribeiro Dos Santos, A.M.; Fernandes, M.R.; Pinto, P.; Wanderley, A.V.; Souza, S.J.D.; Kroll, J.E.; Pereira, A.L.; et al. Identification of NUDT15 Gene Variants in Amazonian Amerindians and Admixed Individuals from Northern Brazil. PLoS ONE 2020, 15, e0231651. [Google Scholar] [CrossRef] [Green Version]

| Gene | SNP ID | Region | Alleles | Impact Predicted by SNPeff | IND | AFR | AMR | EAS | EUR | SAS |
|---|---|---|---|---|---|---|---|---|---|---|
| CHD8 | rs35057134 | Intronic | GA > G | Modifier | 0.0143 | 0.2250 | 0.2070 | 0.3480 | 0.2740 | 0.2550 |
| CHD8 | rs80311097 | Intronic | C > A | Modifier | 0.0000 | 0.0610 | 0.0010 | 0.0000 | 0.0010 | 0.0000 |
| CHD8 | rs10467770 | CDS | C > T | Moderate | 0.0781 | 0.2240 | 0.1900 | 0.3450 | 0.2450 | 0.2490 |
| CHD8 | rs111250264 | CDS | G > A | Moderate | 0.0086 | 0.0050 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| CHD8 | rs57764234 | Intronic | C > T | Modifier | 0.0246 | 0.3160 | 0.0290 | 0.0000 | 0.0210 | 0.0050 |
| CHD8 | rs111776414 | Intronic | G > GA | Modifier | 0.0417 | 0.1610 | 0.0120 | 0.0010 | 0.0020 | 0.0110 |
| CHD8 | rs1998332 | Intronic | G > A | Modifier | 0.6172 | 0.5730 | 0.7780 | 0.8880 | 0.9060 | 0.9170 |
| CHD8 | rs149307240 | CDS | C > T | Moderate | 0.0259 | 0.0000 | 0.0160 | 0.0000 | 0.0010 | 0.0000 |
| SCN2A | rs17183814 | CDS | G > A | Moderate | 0.2500 | 0.0210 | 0.0820 | 0.1380 | 0.0570 | 0.1420 |
| SCN2A | rs75109281 | Intronic | C > T | Modifier | 0.0833 | 0.0120 | 0.0030 | 0.0000 | 0.0000 | 0.0000 |
| SCN2A | rs3769951 | Intronic | C > T | Modifier | 0.0135 | 0.1640 | 0.2480 | 0.2610 | 0.2920 | 0.3310 |
| SCN2A | rs28472553 | Intronic | A > C | Modifier | 0.0833 | 0.0290 | 0.0030 | 0.0000 | 0.0010 | 0.0000 |
| SCN2A | rs139906774 | Intronic | G > GA | Modifier | 0.0000 | 0.0520 | 0.3000 | 0.3420 | 0.2420 | 0.1860 |
| SCN2A | rs2304014 | Intronic | T > A | Modifier | 0.0270 | 0.2280 | 0.1330 | 0.1410 | 0.1760 | 0.1390 |
| SCN2A | rs6432821 | Intronic | T > C | Modifier | 1.0000 | 0.9520 | 0.9970 | 1.0000 | 1.0000 | 0.9990 |
| SCN2A | rs150453735 | Intronic | C > T | Modifier | 0.1852 | 0.0020 | 0.0530 | 0.0000 | 0.0000 | 0.0000 |
| SCN2A | rs1867864 | Intronic | C > T | Modifier | 0.4453 | 0.6130 | 0.4600 | 0.3430 | 0.5640 | 0.4870 |
| SCN2A | rs1838846 | Intronic | A > G | Modifier | 0.0000 | 0.7930 | 0.7940 | 0.7430 | 0.8300 | 0.6980 |
| SCN2A | rs7593568 | Intronic | A > G | Modifier | 0.0000 | 0.7950 | 0.7950 | 0.7430 | 0.8300 | 0.6950 |
| FOXP1 | rs1435680522 | 3UTR | GT > G | Modifier | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| FOXP1 | rs112773801 | 3UTR | G > GT | Modifier | 0.0167 | 0.4240 | 0.1860 | 0.4510 | 0.1340 | 0.3290 |
| FOXP1 | rs58847217 | Intronic | T > C | Modifier | 0.0278 | 0.1010 | 0.0040 | 0.0000 | 0.0000 | 0.0000 |
| FOXP1 | rs76145927 | CDS | T > C | Moderate | 0.0000 | 0.0000 | 0.0060 | 0.0390 | 0.0030 | 0.0000 |
| FOXP1 | rs72960080 | Intronic | T > C | Modifier | 0.0833 | 0.1190 | 0.0040 | 0.0000 | 0.0000 | 0.0000 |
| FOXP1 | rs13068094 | Intronic | C > T | Modifier | 0.0833 | 0.1040 | 0.5400 | 0.0550 | 0.5750 | 0.2820 |
| FOXP1 | rs7638391 | Intronic | G > T | Modifier | 1.0000 | 0.9970 | 0.9650 | 1.0000 | 0.9230 | 0.9780 |
| FOXP1 | rs56850311 | Intronic | A > T | Modifier | 0.0000 | 0.3960 | 0.2520 | 0.1060 | 0.2890 | 0.2230 |
| FOXP1 | rs7639736 | Intronic | C > A | Modifier | 0.0000 | 0.0760 | 0.0560 | 0.0730 | 0.0130 | 0.0200 |
| FOXP1 | rs939845 | Intronic | A > G | Modifier | 0.3984 | 0.1630 | 0.2250 | 0.1110 | 0.0640 | 0.0440 |
| FOXP1 | rs2037474 | Intronic | A > G | Modifier | 0.5156 | 0.2720 | 0.3430 | 0.4360 | 0.1360 | 0.2450 |
| FOXP1 | rs151011253 | Intronic | T > TA | Modifier | 0.0139 | 0.0850 | 0.0560 | 0.0310 | 0.0540 | 0.0960 |
| SYNGAP1 | rs76557362 | Intronic | C > T | Modifier | 0.0833 | 0.2530 | 0.0130 | 0.0000 | 0.0000 | 0.0000 |
| SYNGAP1 | rs453590 | Intronic | C > T | Modifier | 0.0000 | 0.2700 | 0.4060 | 0.6410 | 0.3860 | 0.5430 |
| SYNGAP1 | rs115441992 | Intronic | C > T | Modifier | 0.0833 | 0.0140 | 0.0090 | 0.0000 | 0.0130 | 0.0020 |
| SYNGAP1 | rs9394145 | Intronic | C > T | Modifier | 0.5078 | 0.0130 | 0.3240 | 0.2500 | 0.3150 | 0.3220 |
| Gene | DbSNP | IND vs. AFR * | IND vs. AMR * | IND vs. EAS * | IND vs. EUR * | IND vs. SAS * |
|---|---|---|---|---|---|---|
| CHD8 | rs35057134 | 5.98 × 10−6 | 2.74 × 10−5 | 5.31 × 10−10 | 1.39 × 10−7 | 7.66 × 10−7 |
| CHD8 | rs80311097 | 0.24887 | 0.28751 | 0.21283 | 0.21318 | 0.21826 |
| CHD8 | rs10467770 | 0.00572 | 0.03028 | 2.97 × 10−6 | 0.00220 | 0.00138 |
| CHD8 | rs111250264 | 0.30959 | 0.28751 | 0.21283 | 0.21318 | 0.21826 |
| CHD8 | rs57764234 | 6.46 × 10−8 | 1.00000 | 0.03481 | 0.64915 | 0.06785 |
| CHD8 | rs111776414 | 0.01566 | 0.07929 | 0.00504 | 0.00507 | 0.05415 |
| CHD8 | rs1998332 | 0.50781 | 0.01151 | 4.40 × 10−7 | 3.06 × 10−8 | 6.23 × 10−9 |
| CHD8 | rs149307240 | 0.02173 | 0.36125 | 0.03481 | 0.03493 | 0.03665 |
| SCN2A | rs17183814 | 1.19 × 10−10 | 0.00026 | 0.02585 | 5.70 × 10−6 | 0.04046 |
| SCN2A | rs75109281 | 0.00339 | 0.00042 | 8.63 × 10−5 | 8.70 × 10−5 | 9.84 × 10−5 |
| SCN2A | rs3769951 | 0.00037 | 1.60 × 10−6 | 4.39 × 10−7 | 3.69 × 10−8 | 2.16 × 10−9 |
| SCN2A | rs28472553 | 0.05210 | 0.00042 | 8.63 × 10−5 | 8.70 × 10−5 | 9.84 × 10−5 |
| SCN2A | rs139906774 | 0.35386 | 3.54 × 10−8 | 6.13 × 10−10 | 1.54 × 10−6 | 0.00011 |
| SCN2A | rs2304014 | 3.85 × 10−5 | 0.01857 | 0.00941 | 0.00166 | 0.01459 |
| SCN2A | rs6432821 | 0.10268 | 1.00000 | 1.00000 | 1.00000 | 1.00000 |
| SCN2A | rs150453735 | 9.91 × 10−13 | 0.00066 | 1.86 × 10−11 | 1.90 × 10−11 | 2.57 × 10−11 |
| SCN2A | rs1867864 | 0.01590 | 1.00000 | 0.09628 | 0.10907 | 0.69027 |
| SCN2A | rs1838846 | 8.09 × 10−38 | 3.83 × 10−35 | 5.39 × 10−32 | 1.00 × 10−40 | 2.50 × 10−28 |
| SCN2A | rs7593568 | 5.55 × 10−38 | 3.83 × 10−35 | 5.39 × 10−32 | 1.00 × 10−40 | 3.54 × 10−28 |
| FOXP1 | rs1435680522 | 0.16887 | 0.28751 | 0.21283 | 0.21318 | 0.21826 |
| FOXP1 | rs112773801 | 2.15 × 10−13 | 0.00013 | 3.53 × 10−14 | 0.00344 | 2.18 × 10−9 |
| FOXP1 | rs58847217 | 0.07415 | 0.06453 | 0.03481 | 0.03493 | 0.03665 |
| FOXP1 | rs76145927 | 0.16887 | 0.39900 | 0.49517 | 0.30231 | 0.21827 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa, G.E.; Fernandes, G.L.; Rodrigues, J.C.G.; da V. B. Leal, D.F.; Pastana, L.F.; Pereira, E.E.B.; Assumpção, P.P.; Burbano, R.M.R.; dos Santos, S.E.B.; Guerreiro, J.F.; et al. Exome Evaluation of Autism-Associated Genes in Amazon American Populations. Genes 2022, 13, 368. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13020368
da Costa GE, Fernandes GL, Rodrigues JCG, da V. B. Leal DF, Pastana LF, Pereira EEB, Assumpção PP, Burbano RMR, dos Santos SEB, Guerreiro JF, et al. Exome Evaluation of Autism-Associated Genes in Amazon American Populations. Genes. 2022; 13(2):368. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13020368
Chicago/Turabian Styleda Costa, Giovana E., Giordane L. Fernandes, Juliana C. G. Rodrigues, Diana F. da V. B. Leal, Lucas F. Pastana, Esdras E. B. Pereira, Paulo P. Assumpção, Rommel M. R. Burbano, Sidney E. B. dos Santos, João F. Guerreiro, and et al. 2022. "Exome Evaluation of Autism-Associated Genes in Amazon American Populations" Genes 13, no. 2: 368. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13020368