Genetic Dissection of Azuki Bean Weevil (Callosobruchus chinensis L.) Resistance in Moth Bean (Vigna aconitifolia [Jaqc.] Maréchal)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Evaluation of Seeds for Bruchid Resistance
2.3. Correlation Analysis and Determination of the Mode of Inheritance of Resistance
2.4. Estimation of the Heritability of Resistance
2.5. Quantitative Trait Loci Analysis
2.6. Comparative Genomic Analysis of Bruchid-Resistance QTLs in Moth Bean, Mungbean and Wild Azuki Bean Relatives
3. Results
3.1. Variation in Callosobruchus chinensis Resistance in Parents and F2 and F2:3 Generations
3.2. Correlations among Traits
3.3. Segregation Analysis and Heritability of Resistance
3.4. QTL Analysis
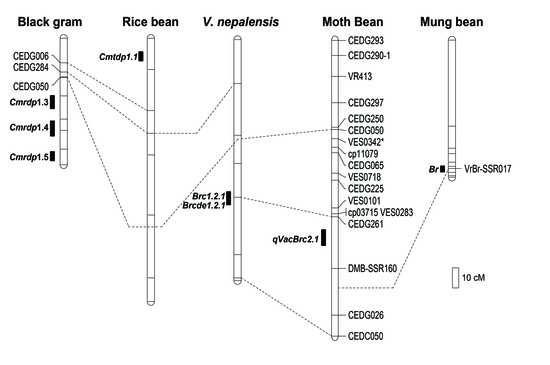
3.5. Comparison of QTLs for Bruchid Resistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rees, D.P. Insects of Stored Products; CSIRO Publishing: Melbourne, Australia, 2004; pp. 39–46. [Google Scholar]
- Talekar, N.S. Biology, damage and control of bruchid pests of mungbean. In Proceedings of the 2nd International Symposium on Mungbean, Bangkok, Thailand, 16–20 November 1987; Shanmugasundaram, S., McLean, B.T., Eds.; AVRDC: Tainan, Taiwan, 1988; pp. 329–342. [Google Scholar]
- Srinives, P.; Somta, P.; Somta, C. Genetics and breeding of resistance to bruchids (Callosobruchus spp.) in Vigna crops: A review. NU. Int. J. Sci. 2007, 4, 1–17. [Google Scholar]
- Singh, S.R. Cowpea cultivars resistant to insect pests in world germplasm collection. Trop. Grain Legume Bull. 1977, 9, 1–7. [Google Scholar]
- Fujii, K.; Miyazaki, S. Infestation resistance of wild legumes (Vigna sublobata) to azuki bean weevil, Callosobruchus chinensis (L.) (Coleoptera: Bruchidae) and its relationship with cytogenetic classification. Appl. Entomol. Zool. 1987, 22, 319–322. [Google Scholar] [CrossRef]
- Talekar, N.S.; Lin, C.P. Characterization of Callosobruchus chinensis (Coleoptera: Bruchidae) resistance in mungbean. J. Econ. Entomol. 1992, 85, 1150–1153. [Google Scholar] [CrossRef]
- Lambrides, C.J.; Imrie, B.C. Susceptibility of mungbean varieties to the bruchid species Callosobruchus maculatus (F.), C phaseoli (Gyll.), C chinensis (L.), and Acanthoscelides obtectus (Say) (Coleoptera: Chrysomelidae). Aust. J. Agric. Res. 2000, 51, 85–89. [Google Scholar] [CrossRef]
- Somta, C.; Somta, P.; Tomooka, N.; Ooi, P.A.C.; Vaughan, D.A.; Srinives, P. Characterization of new sources of mungbean (Vigna radiata (L.) Wilczek) resistance to bruchids., Callosobruchus spp. (Coleoptera: Bruchidae). J. Stored Prod. Res. 2008, 44, 316–321. [Google Scholar] [CrossRef]
- Vaughan, D.A.; Tomooka, N.; Kaga, A. Adzuki bean (Vigna angularis (Willd.) Ohwi & Ohashi). In Genetic Resources, Chromosome Engineering, and Crop Improvement; Singh, R.J., Pauhar, P.P., Eds.; Taylor and Francis Group LLC: Boca Raton, FL, USA, 2005; pp. 347–359. [Google Scholar]
- Somta, P.; Kaga, A.; Tomooka, N.; Isemura, T.; Vaughan, D.A.; Srinives, P. Mapping of quantitative trait loci for a new source of resistance to bruchids in the wild species Vigna nepalensis Tateishi and Maxted (Vigna subgenus Ceratotropis). Theor. Appl. Genet. 2008, 117, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Blink, M.; Jansen, P.C.M. Vigna aconitifolia (Jaqc.) Maréchal. In Plant Resources of Tropical Africa 1: Cereals and Pulses; Blink, M., Belay, G., Eds.; PROTA Foundation/Backhuys Publisher/CTA: Wageningen, The Netherlands, 2006; pp. 200–203. [Google Scholar]
- Van Oers, C.C.C.M. Vigna aconitifolia (Jaqc.) Maréchal. In Plant Resources of South-East Asia 1: Pulses; Maesen, V.D., Somaatmadja, S., Eds.; PROSEA Foundation: Bogor, Indonesia, 1992; pp. 66–67. [Google Scholar]
- Kumar, D. Production Technology for Moth Bean in India; Indian Turisum Publication: Jodhpur, India, 2002; pp. 1–29. [Google Scholar]
- Tomooka, N.; Kashiwaba, K.; Vaughan, D.A.; Ishimoto, M.; Egawa, Y. The effectiveness of evaluating wild species: Searching for sources of resistance to bruchids beetles in the genus Vigna subgenus Ceratotropis. Euphytica 2000, 115, 27–41. [Google Scholar] [CrossRef]
- Yundaeng, C.; Somta, P.; Amkul, K.; Kongjaimun, K.; Kaga, A.; Pandiyan, M.; Natesan, S.; Tomooka, N. Construction of genetic linkage map and genome dissection of domestication related traits of moth bean (Vigna aconitifolia), a legume crop of arid areas. Mol. Genet. Genom. 2018. submitted. [Google Scholar]
- Somta, P.; Ammaranan, C.; Ooi, P.A.C.; Srinives, P. Inheritance of seed resistance to bruchids in cultivated mungbean (Vigna radiata L. Wilczek). Euphytica 2007, 155, 47–55. [Google Scholar] [CrossRef]
- Van der Plank, J.E. Plant Diseases: Epidemics and Control; Academic Press: New York, NY, USA, 1963; pp. 1–349. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Li, H.; Ye, G.; Wang, J. A modified algorithm for the improvement of composite interval mapping. Genetics 2007, 175, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Chotechung, S.; Somta, P.; Chen, J.; Yimram, T.; Chen, X.; Srinives, P. A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 2016, 129, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Kim, S.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J.; et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakai, H.; Naito, K.; Ogiso-Tanaka, E.; Takahashi, Y.; Iseki, K.; Muto, C.; Satou, K.; Teruya, K.; Shiroma, A.; Shimoji, M.; et al. The power of single molecule real-time sequencing technology in the de novo assembly of a eukaryotic genome. Sci Rep. 2015, 5, 16780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkataramana, P.B.; Gowda, R.; Somta, P.; Ramesh, S.; Rao, A.M.; Bhanuprakash, K.; Srinives, P.; Gireesh, C.; Pramila, C.K. Mapping QTL for bruchid resistance in rice bean (Vigna umbellata). Euphytica 2016, 207, 135–147. [Google Scholar] [CrossRef]
- Kaewwongwal, A.; Chen, J.; Somta, P.; Kongjaimun, A.; Yimram, T.; Chen, X.; Srinives, P. Novel alleles of two tightly linked gene encoding polygalacturonase-inhibiting proteins (VrPGIP1 and VrPGIP2) associated with the Br locus that confer bruchid (Callosobruchus spp.) resistance to mungbean (Vigna radiata) accession V2709. Front. Plant Sci. 2017, 8, 1692. [Google Scholar] [CrossRef] [PubMed]
- Souframanien, J.; Gupta, S.K.; Gopalakrishna, T. Identification of quantitative trait loci for bruchid (Callosobruchus maculatus) resistance in black gram [Vigna mungo (L.) Hepper]. Euphytica 2010, 176, 349–356. [Google Scholar] [CrossRef]
- Kaga, A.; Isemura, T.; Shimizu, T.; Somta, P.; Srinives, P.; Tabata, S.; Tomooa, N.; Vaughan, D.A. Asian Vigna genome research. In Proceedings of the 14th NIAS International Workshop on Genetic Resources: Genetic Resources and Comparative Genomics of Legumes (Glycine and Vigna), Tsukuba, Japan, 14 September 2009; Tomooka, N., Vaughan, D.A., Eds.; National Institute of Agrobiological Sciences: Tsukuba, Japan, 2011; pp. 33–39. [Google Scholar]
- Kitamura, K.; Ishimoto, M.; Sawa, M. Inheritance of resistance to infestation with azuki bean weevil in Vigna sublobata and successful incorporation to V. radiata. Jpn. J. Breed. 1988, 38, 459–464. [Google Scholar] [CrossRef]
- Dongre, T.K.; Pawar, S.E.; Thakare, R.G.; Harwalkar, M.R. Identification of resistant source to cowpea weevil (Callosobruchus maculatus (F.)) in Vigna sp. and inheritance of their resistance in black gram (Vigna mungo var. mungo). J. Stored Prod. Res. 1996, 32, 201–204. [Google Scholar] [CrossRef]
- Redden, R.J.; McGuire, J. The genetic evaluation of bruchid resistance in seed of cowpea. Aust. J. Agric. Res. 1983, 34, 707–715. [Google Scholar] [CrossRef]
- Mei, L.; Cheng, X.Z.; Wang, S.H.; Wang, L.X.; Liu, C.Y.; Sun, L.; Xu, N.; Humphry, M.E.; Lambrides, C.J.; Li, H.B.; et al. Relationship between bruchid resistance and seed mass in mungbean based on QTL analysis. Genome 2009, 52, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Somta, P.; Kaga, A.; Tomooka, N.; Kashiwaba, K.; Isemura, K.; Chaitieng, B.; Srinives, P.; Vaughan, D.A. Development of an interspecific Vigna linkage map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant Breed. 2006, 125, 77–84. [Google Scholar] [CrossRef]
- Isemura, T.; Kaga, A.; Konishi, S.; Ando, T.; Tomooka, N.; Han, O.K.; Vaughan, D.A. Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm season legumes. Ann. Bot. 2007, 100, 1053–1071. [Google Scholar] [CrossRef] [PubMed]
- Isemura, T.; Kaga, A.; Tomooka, N.; Shimizu, T.; Vaughan, D.A. The genetics of domestication of rice bean, Vigna umbellata. Ann. Bot. 2010, 106, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Isemura, T.; Kaga, A.; Tabata, S.; Somta, P.; Srinives, P.; Shimizu, T.; Jo, U.; Vaughan, D.A.; Tomooka, N. Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 2012, 7, e41304. [Google Scholar] [CrossRef] [PubMed]
- Kongjaimun, A.; Kaga, A.; Tomooka, N.; Somta, P.; Shimizu, T.; Shu, Y.; Isemura, T.; Vaughan, D.A.; Srinives, P. An SSR-based linkage map of yardlong bean (Vigna unguiculata ssp. sesquipedalis (L.) Verc.) and QTL analysis of pod length. Genome 2012, 55, 81–92. [Google Scholar] [CrossRef]






| Population | Percentage of Damaged Seeds | AUDPC | ||||
|---|---|---|---|---|---|---|
| Min–Max | Mean | Heritability (%) | Min–Max | Mean | Heritability (%) | |
| IPCMO056 | 95.4–100 | 98.1 | - | 2826.0–3158.0 | 2992.0 | - |
| TN67 | 6.6–21.6 | 11.9 | - | 197.0–500.0 | 348.5 | - |
| F2 population | 0.0–100 | 41.6 | 83.60 | 0.0–3103.1 | 1163.2 | 85.99 |
| F2:3 population (2016) | 3.5–99.6 | 57.0 | 90.42 | Not determined | ||
| F2:3 population (2017) (Set I) | 0.5–100 | 53.1 | 96.03 | 16.4–3385.0 | 1536.7 | 98.65 |
| F2:3 population (2017) (Set II) | 1.0–100 | 52.2 | 99.97 | Not determined | ||
| Population | Trait | No. of Plants/Lines Tested | Resistant: Susceptible | Chi-Square (p Value) |
|---|---|---|---|---|
| F2 | % damaged seeds | 187 | 143:44 | 0.2157 (0.6423) |
| AUDPC | 187 | 142:45 | 0.0873 (0.7676) | |
| F2:3-A 1 | % damaged seeds | 172 | 125:47 | 0.4961 (0.4812) |
| F2:3-B2 (set I) | % damaged seeds | 166 | 121:45 | 0.3936 (0.5304) |
| F2:3-B2 (set II) | % damaged seeds | 166 | 122:44 | 0.2880 (0.6541) |
| AUDPC | 166 | 116:50 | 2.3213 (0.1276) |
| Population | Trait | LG a | QTL name | Position b | Franking Markers | LOD | PVE c (%) | Add d | Dom e |
|---|---|---|---|---|---|---|---|---|---|
| F2 | % damaged seeds | 2 | qVacPDS2.1 | 91 | CEDG261—DMB-SSR160 | 69.55 | 62.98 | 42.64 | −11.16 |
| AUDPC | 2 | qVacAUDPC2.1 | 91 | CEDG261—DMB-SSR160 | 82.20 | 63.00 | 1273.19 | −483.39 | |
| 100-seed weight | 3 | qVacSDW3.1 | 22 | VES084—CEDG155 | 3.72 | 6.48 | 0.10 | 0.01 | |
| 4 | qVacSDW4.2 | 65 | CEDG091—CEDG165 | 6.38 | 11.11 | 0.14 | −0.02 | ||
| 5 | qVacSDW5.1 | 0 | CEDG020—VES0091 | 5.28 | 9.01 | 0.11 | −0.03 | ||
| 6 | qVacSDW6.2 | 71 | CEDG146—cp09781 | 7.25 | 15.56 | 0.15 | −0.02 | ||
| F2:3-A | % damaged seeds | 2 | qVacPDS2.1 | 90 | CEDG261—DMB-SSR160 | 40.22 | 50.41 | 32.91 | 2.98 |
| 5 | qVacPDS5.2 | 17 | CEDG264—VES0664 | 14.54 | 12.19 | 0.51 | 22.76 | ||
| 100-seed weight | 2 | qVacSDW2.1 | 36 | CDEG297—CEDG250 | 4.66 | 7.93 | 0.09 | −0.01 | |
| 3 | qVacSDW3.2 | 45 | VES0053—CEDG084 | 7.49 | 11.82 | 0.09 | 0.04 | ||
| 4 | qVacSDW4.1 | 32 | Bms—VES0675 | 14.01 | 26.84 | 0.14 | −0.03 | ||
| 5 | qVacSDW5.2 | 18 | VES0664—CEDG027 | 4.86 | 7.66 | 0.07 | 0.03 | ||
| 6 | qVacSDW6.1 | 20 | CEDG169—CEDG034 | 3.65 | 5.80 | 0.67 | 0.01 | ||
| F2:3-B (Set I) | % damaged seeds | 2 | qVacPDS2.1 | 91 | CEDG261—DMB-SSR160 | 46.79 | 61.32 | 41.42 | 0.73 |
| AUDPC | 2 | qVacAUDPC2.1 | 91 | CEDG261—DMB-SSR160 | 44.90 | 58.73 | 1279.74 | −90.70 | |
| 100-seed weight | 1 | qVacSDW1.1 | 2 | CEDG149—CEDC007 | 4.51 | 6.75 | 0.08 | 0.03 | |
| 3 | qVacSDW3.2 | 55 | VR169—VES0070 | 6.98 | 14.01 | 0.11 | 0.01 | ||
| 4 | qVacSDW4.1 | 28 | Bms—VES0675 | 8.43 | 16.03 | 0.11 | −0.05 | ||
| 4 | qVacSDW4.2 | 63 | CEDG091—CEDG165 | 4.22 | 6.24 | 0.07 | −0.01 | ||
| 5 | qVacSDW5.1 | 0 | CEDG020—CEDG091 | 3.61 | 4.80 | 0.06 | −0.01 | ||
| 7 | qVacSDW7.1 | 19 | CEDG174—CEDG215 | 5.23 | 8.46 | 0.83 | 0.01 | ||
| F2:3-B (Set II) | % damaged seeds | 2 | qVacPDS2.1 | 91 | CEDG261—DMB-SSR160 | 47.97 | 64.23 | 40.98 | 0.10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Somta, P.; Jomsangawong, A.; Yundaeng, C.; Yuan, X.; Chen, J.; Tomooka, N.; Chen, X. Genetic Dissection of Azuki Bean Weevil (Callosobruchus chinensis L.) Resistance in Moth Bean (Vigna aconitifolia [Jaqc.] Maréchal). Genes 2018, 9, 555. https://0-doi-org.brum.beds.ac.uk/10.3390/genes9110555
Somta P, Jomsangawong A, Yundaeng C, Yuan X, Chen J, Tomooka N, Chen X. Genetic Dissection of Azuki Bean Weevil (Callosobruchus chinensis L.) Resistance in Moth Bean (Vigna aconitifolia [Jaqc.] Maréchal). Genes. 2018; 9(11):555. https://0-doi-org.brum.beds.ac.uk/10.3390/genes9110555
Chicago/Turabian StyleSomta, Prakit, Achara Jomsangawong, Chutintorn Yundaeng, Xingxing Yuan, Jingbin Chen, Norihiko Tomooka, and Xin Chen. 2018. "Genetic Dissection of Azuki Bean Weevil (Callosobruchus chinensis L.) Resistance in Moth Bean (Vigna aconitifolia [Jaqc.] Maréchal)" Genes 9, no. 11: 555. https://0-doi-org.brum.beds.ac.uk/10.3390/genes9110555