Use of Advanced Artificial Intelligence in Forensic Medicine, Forensic Anthropology and Clinical Anatomy
Abstract
:1. Introduction
1.1. Basic Terminology and Principles in Era of AI Enhanced Forensic Medicine
1.2. Overview of Used Artificial Intelligence for Forensic Age and Sex Determination
1.3. Artificial Intelligence Implementation in 3D Cephalometric Landmark Identification
1.4. Artificial Intelligence Implementation in Soft-Tissue Face Prediction from Skull and Vice Versa
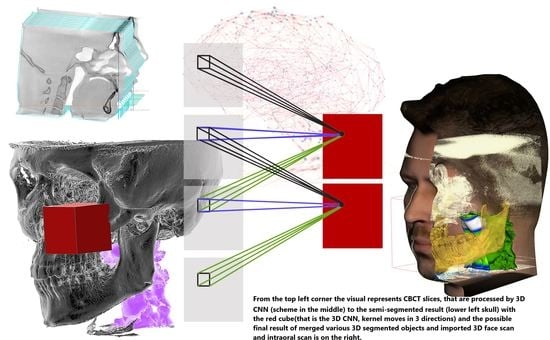
2. 3D Convolutional Neural Networks and Methods of Their Use in Forensic Medicine
2.1. Hardware and Software Used
2.2. Main Tasks Definitions
2.3. The Data Management
- Loading DICOM files—the proper way of loading the DICOM file ensures that we will not lose the exact quality
- Pixel values to Hounsfield Units alignment—the Hounsfield Unit (HU) measures radiodensity for each body tissue. The Hounsfield scale that determines the values for various tissues usually ranges from −1000 HU to +3000 HU, and therefore, this step ensures that the pixel values for each CT scan do not exceed these thresholds.
- Resampling to isomorphic resolution—the distance between consecutive slices in each CT scan defines the slice thickness. This would mean a nontrivial challenge for the neural network. The thickness depends on the CT device setup, and therefore it is necessary to create equally spaced slices.
- [Optional] Specific part segmentation—each tissue corresponds to a specific range in the Hounsfield units scale, and in some cases, we can segment out specific parts of the CT scan by thresholding the image.
- Normalization and zero centering—these two steps ensure that the input data that are feed into the neural network are normalized into [0, 1] interval (normalization) and are zero centered (achieved by subtracting the mean value of the image pixel values).
2.4. Dataset Specification
2.5. Deep Learning Approach
2.5.1. Age Estimation Using 3D Deep Neural Networks
2.5.2. Sex Classification Using 3D Deep Neural Networks
2.5.3. Automatization of Cephalometric Analysis
- Landmarks estimation in whole CT scan image—in this approach, the probability estimation for all landmarks is assigned for each pixel in the CT scan
- Landmarks estimation for selected regions of interest—assuming that each landmark corresponds to a specific area we could add another preprocessing step—slice cut where each slice would be a template-based region fed into a neural network. We can determine the expected landmark detection for each slice independently, which should help in the final model performance
2.5.4. Neural Networks Architectures and Clinical Data Pre-Processing
- Use whole 3D CT scan as an input into the neural network and output one value for age estimation as floating value and one for sex classification as a binary value.
- Segment out the mandible and use it as input into the neural network. Output is the same as in the previous task.
- (experimental) Use a whole 3D CT scan to input into the neural network and output multiple values representing specific skull features (as discussed at the meeting last week). Then use these values as an input into another machine learning model to estimate age and gender.
2.6. Evaluation
2.6.1. Regression Models
2.6.2. Classification Models
3. Resulting Summary of Proposed Approach for Utilization of 3D CNN in Investigated Aspects of Forensic Medicine
4. Discussion
- Biological age determination
- Sex determination
- Automatized, precise and reliable:
- ○
- 3D cephalometric analysis of soft and hard tissues
- ○
- 3D face prediction from the skull (soft-tissues) and vice versa
- ○
- Search for hidden damage in post-mortem high-resolution CT images
- ○
- Asymmetry and disproportionality evaluation
- Predictions of:
- ○
- Hard-tissue and soft tissue growth
- ○
- Aging in general
- ○
- Ideal face proportions respecting golden ratio proportions
- 3D reconstructions of:
- ○
- Missing parts of the skull or face
- ○
- 3D dental fingerprints for identification with 2D dental records
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D CNN | Three-Dimensional Convolutional Neural Network |
| AI | Artificial Intelligence |
| CBCT | Cone-Beam Computer Tomography |
| CM | Confusion Matrix |
| DICOM | Communications in Medicine Format |
| DA | Data Augmentation |
| FOV | Field of View |
| HU | Hounsfield Unit |
| ML | Machine Learning |
| MAE | Mean Absolute Error |
| MSE | Mean Squared Error |
| nMR | Nuclear Magnetic Resonance |
| STC | Slice Timing Correction |
References
- Ya-ting, F.A.; Qiong, L.A.; Tong, X.I. New Opportunities and Challenges for Forensic Medicine in the Era of Artificial Intelligence Technology. J. Forensic Med. 2020, 36, 77. [Google Scholar] [CrossRef]
- Khanagar, S.B.; Vishwanathaiah, S.; Naik, S.; Al-Kheraif, A.A.; Devang Divakar, D.; Sarode, S.C.; Bhandi, S.; Patil, S. Application and Performance of Artificial Intelligence Technology in Forensic Odontology—A Systematic Review. Leg. Med. 2021, 48, 101826. [Google Scholar] [CrossRef]
- Lefèvre, T. Big Data in Forensic Science and Medicine. J. Forensic Leg. Med. 2018, 57, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pathak, M.; Narang, H. Application of Artificial Intelligence in the Field of Forensic Medicine. Indian J. Forensic Med. Toxicol. 2021, 15. [Google Scholar] [CrossRef]
- Franke, K.; Srihari, S.N. Computational Forensics: An Overview. In Computational Forensics; Springer: Berlin/Heidelberg, Germany, 2008; Volume 5158, pp. 1–10. [Google Scholar] [CrossRef]
- Cossellu, G.; de Luca, S.; Biagi, R.; Farronato, G.; Cingolani, M.; Ferrante, L.; Cameriere, R. Reliability of Frontal Sinus by Cone Beam-Computed Tomography (CBCT) for Individual Identification. Radiol. Med. 2015, 120, 1130–1136. [Google Scholar] [CrossRef]
- der Mauer, M.A.; Well, E.J.; Herrmann, J.; Groth, M.; Morlock, M.M.; Maas, R.; Säring, D. Automated Age Estimation of Young Individuals Based on 3D Knee MRI Using Deep Learning. Int. J. Leg. Med. 2020, 135, 649–663. [Google Scholar] [CrossRef]
- Armanious, K.; Abdulatif, S.; Bhaktharaguttu, A.R.; Küstner, T.; Hepp, T.; Gatidis, S.; Yang, B. Organ-Based Chronological Age Estimation Based on 3D MRI Scans. In Proceedings of the 28th European Signal Processing Conference, Amsterdam, The Netherlands, 24–28 August 2021; pp. 1225–1228. [Google Scholar] [CrossRef]
- Souadih, K.; Belaid, A.; Salem, D. Automatic Segmentation of the Sphenoid Sinus in CT-Scans Volume with DeepMedics 3D CNN Architecture. Med. Technol. J. 2019, 3, 334–346. [Google Scholar] [CrossRef]
- Dubey, C.; Raj, S.; Munuswamy, S.; Katta, A.; Chetty, G. Semantic Segmentation Using Deep Neural Networks in Medicine—A Survey. ISO 2018, 3, 39–48. [Google Scholar]
- Du, R.; Vardhanabhuti, V. 3D-RADNet: Extracting Labels from DICOM Metadata for Training General Medical Domain Deep 3D Convolution Neural Networks. Proc. Mach. Learn. Res. 2020, 121, 174–192. [Google Scholar]
- Fajar, A.; Sarno, R.; Fatichah, C.; Fahmi, A. Reconstructing and Resizing 3D Images from DICOM Files. J. King Saud Univ. Comput. Inf. Sci. 2020. [Google Scholar] [CrossRef]
- da Silva, R.D.C.; Jenkyn, T.; Carranza, V. Convolutional Neural Networks and Geometric Moments to Identify the Bilateral Symmetric Midplane in Facial Skeletons from CT Scans. Biology 2021, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D Deep Learning on Medical Images: A Review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef]
- Armanious, K.; Abdulatif, S.; Shi, W.; Salian, S.; Küstner, T.; Weiskopf, D.; Hepp, T.; Gatidis, S.; Yang, B. Age-Net: An MRI-Based Iterative Framework for Biological Age Estimation. arXiv 2009, arXiv:10765.14-37. [Google Scholar] [CrossRef]
- Sajedi, H.; Pardakhti, N. Age Prediction Based on Brain MRI Image: A Survey. J. Med Syst. 2019, 43, 1–30. [Google Scholar] [CrossRef]
- de Luca, S.; Alemán, I.; Bertoldi, F.; Ferrante, L.; Mastrangelo, P.; Cingolani, M.; Cameriere, R. Age Estimation by Tooth/Pulp Ratio in Canines by Peri-Apical X-Rays: Reliability in Age Determination of Spanish and Italian Medieval Skeletal Remains. J. Archaeol. Sci. 2010, 37, 3048–3058. [Google Scholar] [CrossRef] [Green Version]
- Cameriere, R.; Ferrante, L.; Cingolani, M. Age Estimation in Children by Measurement of Open Apices in Teeth. Int. J. Leg. Med. 2006, 120, 49–52. [Google Scholar] [CrossRef] [Green Version]
- Hostiuc, S.; Diaconescu, I.; Rusu, M.C.; Negoi, I. Age Estimation Using the Cameriere Methods of Open Apices: A Meta-Analysis. Healthcare 2021, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Cameriere, R.; de Luca, S.; Egidi, N.; Bacaloni, M.; Maponi, P.; Ferrante, L.; Cingolani, M. Automatic Age Estimation in Adults by Analysis of Canine Pulp/Tooth Ratio: Preliminary Results. J. Forensic Radiol. Imaging 2015, 3, 61–66. [Google Scholar] [CrossRef]
- Rai, B.; Kaur, J.; Cingolani, M.; Ferrante, L.; Cameriere, R. Age Estimation in Children by Measurement of Open Apices in Teeth: An Indian Formula. Int. J. Leg. Med. 2010, 124, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Cameriere, R.; Ferrante, L. Canine Pulp Ratios in Estimating Pensionable Age in Subjects with Questionable Documents of Identification. Forensic Sci. Int. 2011, 206, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Cameriere, R.; Giuliodori, A.; Zampi, M.; Galić, I.; Cingolani, M.; Pagliara, F.; Ferrante, L. Age Estimation in Children and Young Adolescents for Forensic Purposes Using Fourth Cervical Vertebra (C4). Int. J. Leg. Med. 2015, 129, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Cameriere, R.; Pacifici, A.; Pacifici, L.; Polimeni, A.; Federici, F.; Cingolani, M.; Ferrante, L. Age Estimation in Children by Measurement of Open Apices in Teeth with Bayesian Calibration Approach. Forensic Sci. Int. 2016, 258, 50–54. [Google Scholar] [CrossRef] [PubMed]
- de Micco, F.; Martino, F.; Velandia Palacio, L.A.; Cingolani, M.; Campobasso, C. pietro Third Molar Maturity Index and Legal Age in Different Ethnic Populations: Accuracy of Cameriere’s Method. Med. Sci. Law 2021, 61, 105–112. [Google Scholar] [CrossRef]
- Scendoni, R.; Cingolani, M.; Giovagnoni, A.; Fogante, M.; Fedeli, P.; Pigolkin, Y.I.; Ferrante, L.; Cameriere, R. Analysis of Carpal Bones on MR Images for Age Estimation: First Results of a New Forensic Approach. Forensic Sci. Int. 2020, 313. [Google Scholar] [CrossRef]
- Cameriere, R.; Santoro, V.; Roca, R.; Lozito, P.; Introna, F.; Cingolani, M.; Galić, I.; Ferrante, L. Assessment of Legal Adult Age of 18 by Measurement of Open Apices of the Third Molars: Study on the Albanian Sample. Forensic Sci. Int. 2014, 245, 205.e1–205.e5. [Google Scholar] [CrossRef]
- de Luca, S.; Mangiulli, T.; Merelli, V.; Conforti, F.; Velandia Palacio, L.A.; Agostini, S.; Spinas, E.; Cameriere, R. A New Formula for Assessing Skeletal Age in Growing Infants and Children by Measuring Carpals and Epiphyses of Radio and Ulna. J. Forensic Leg. Med. 2016, 39, 109–116. [Google Scholar] [CrossRef]
- Cameriere, R.; de Luca, S.; Biagi, R.; Cingolani, M.; Farronato, G.; Ferrante, L. Accuracy of Three Age Estimation Methods in Children by Measurements of Developing Teeth and Carpals and Epiphyses of the Ulna and Radius. J. Forensic Sci. 2012, 57, 1263–1270. [Google Scholar] [CrossRef]
- Cameriere, R.; de Luca, S.; de Angelis, D.; Merelli, V.; Giuliodori, A.; Cingolani, M.; Cattaneo, C.; Ferrante, L. Reliability of Schmeling’s Stages of Ossification of Medial Clavicular Epiphyses and Its Validity to Assess 18 Years of Age in Living Subjects. Int. J. Leg. Med. 2012, 126, 923–932. [Google Scholar] [CrossRef]
- Cameriere, R.; de Luca, S.; Cingolani, M.; Ferrante, L. Measurements of Developing Teeth, and Carpals and Epiphyses of the Ulna and Radius for Assessing New Cut-Offs at the Age Thresholds of 10, 11, 12, 13 and 14 Years. J. Forensic Leg. Med. 2015, 34, 50–54. [Google Scholar] [CrossRef]
- Afrianty, I.; Nasien, D.; Kadir, M.R.A.; Haron, H. Backpropagation Neural Network for Sex Determination from Patella in Forensic Anthropology. In Advances in Computer Science and its Applications; Springer: Berlin/Heidelberg, Germany, 2014; Volume 279, pp. 723–728. [Google Scholar] [CrossRef]
- Pötsch-Schneider, L.; Endris, R.; Schmidt, H. Discriminant Analysis of the Mandible for Sex Determination. Z. Fur Rechtsmedizin. J. Leg. Med. 1985, 94, 21–30. [Google Scholar] [CrossRef]
- el Morsi, D.A.; Gaballah, G.; Tawfik, M.W. Sex Determination in Egyptian Population from Scapula by Computed Tomography. J Forensic Res 2017, 8, 376. [Google Scholar] [CrossRef]
- Kalmey, J.; Rathbun, T. Sex Determination by Discriminant Function Analysis of the Petrous Portion of the Temporal Bone. J. Forensic Sci. 1996, 41, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.; Grajeda, C.; Baggili, I.; Hall, C. A Practitioner Survey Exploring the Value of Forensic Tools, AI, Filtering, & Safer Presentation for Investigating Child Sexual Abuse Material (CSAM). Digit. Investig. 2019, 29, S124–S142. [Google Scholar] [CrossRef]
- Teixeira, W.R. Sex Identification Utilizing the Size of the Foramen Magnum. Am. J. Forensic Med. Pathol. 1982, 3, 203–206. [Google Scholar] [CrossRef]
- Randhawa, K.; Narang, R.S.; Arora, P.C. Study of the effect of age changes on lip print pattern and its reliability in sex determination. J. Forensic Odonto-Stomatol. 2011, 29, 45. [Google Scholar]
- Bewes, J.; Low, A.; Morphett, A.; Pate, F.D.; Henneberg, M. Artificial Intelligence for Sex Determination of Skeletal Remains: Application of a Deep Learning Artificial Neural Network to Human Skulls. J. Forensic Leg. Med. 2019, 62, 40–43. [Google Scholar] [CrossRef]
- Regina Silva Lucena dos Santos, E.; Paulo Feitosa de Albuquerque, P.; Virgínio de Albuquerque, P.; Duarte Ribeiro de Oliveira, B.; Caiaffo, V. Determination of Sex Based on the Morphometric Evaluation of the Proximal Tibia Determinación Del Sexo Basada En La Evaluación Morfométrica de La Tibia Proximal. Int. J. Morphol 2018, 36, 104–108. [Google Scholar] [CrossRef]
- Hwang, H.-W.; Park, J.-H.; Moon, J.-H.; Yu, Y.; Kim, H.; Her, S.-B.; Srinivasan, G.; Aljanabi, M.N.A.; Donatelli, R.E.; Lee, S.-J. Automated Identification of Cephalometric Landmarks: Part 2-Might It Be Better Than Human? Angle Orthod. 2020, 90, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.; Kim, H.P.; Jeon, K.; Lee, S.-H.; Seo, J.K. Automatic 3D Cephalometric Annotation System Using Shadowed 2D Image-Based Machine Learning. Phys. Med. Biol. 2019, 64, 055002. [Google Scholar] [CrossRef] [PubMed]
- Montúfar, J.; Romero, M.; Scougall-Vilchis, R.J. Hybrid Approach for Automatic Cephalometric Landmark Annotation on Cone-Beam Computed Tomography Volumes. Am. J. Orthod. Dentofac. Orthop. 2018, 154, 140–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montúfar, J.; Romero, M.; Scougall-Vilchis, R.J. Automatic 3-Dimensional Cephalometric Landmarking Based on Active Shape Models in Related Projections. Am. J. Orthod. Dentofac. Orthop. 2018, 153, 449–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codari, M.; Caffini, M.; Tartaglia, G.M.; Sforza, C.; Baselli, G. Computer-Aided Cephalometric Landmark Annotation for CBCT Data. Int. J. Comput. Assist. Radiol. Surg. 2016, 12, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Baksi, S.; Freezer, S.; Matsumoto, T.; Dreyer, C. Accuracy of an Automated Method of 3D Soft Tissue Landmark Detection. Eur. J. Orthod. 2020. [Google Scholar] [CrossRef]
- Ed-Dhahraouy, M.; Riri, H.; Ezzahmouly, M.; Bourzgui, F.; el Moutaoukkil, A. A New Methodology for Automatic Detection of Reference Points in 3D Cephalometry: A Pilot Study. Int. Orthod. 2018, 16, 328–337. [Google Scholar] [CrossRef]
- Croquet, B.; Matthews, H.; Mertens, J.; Fan, Y.; Nauwelaers, N.; Mahdi, S.; Hoskens, H.; el Sergani, A.; Xu, T.; Vandermeulen, D.; et al. Automated Landmarking for Palatal Shape Analysis Using Geometric Deep Learning. Orthod. Craniofac. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Lachinov, D.; Getmanskaya, A.; Turlapov, V. Cephalometric Landmark Regression with Convolutional Neural Networks on 3D Computed Tomography Data. Pattern Recognit. Image Anal. 2020, 30, 512–522. [Google Scholar] [CrossRef]
- Dot, G.; Rafflenbeul, F.; Arbotto, M.; Gajny, L.; Rouch, P.; Schouman, T. Accuracy and Reliability of Automatic Three-Dimensional Cephalometric Landmarking. Int. J. Oral Maxillofac. Surg. 2020, 49, 1367–1378. [Google Scholar] [CrossRef]
- Bermejo, E.; Taniguchi, K.; Ogawa, Y.; Martos, R.; Valsecchi, A.; Mesejo, P.; Ibáñez, O.; Imaizumi, K. Automatic Landmark Annotation in 3D Surface Scans of Skulls: Methodological Proposal and Reliability Study. Comput. Methods Programs Biomed. 2021, 210, 106380. [Google Scholar] [CrossRef]
- Lee, J.-H.; Yu, H.-J.; Kim, M.; Kim, J.-W.; Choi, J. Automated Cephalometric Landmark Detection with Confidence Regions Using Bayesian Convolutional Neural Networks. BMC Oral Health 2020, 20, 270. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Jeon, K.; Kang, S.-H.; Lee, S.-H. 3D Cephalometric Landmark Detection by Multiple Stage Deep Reinforcement Learning. Sci. Rep. 2021, 11, 17509. [Google Scholar] [CrossRef] [PubMed]
- Juneja, M.; Garg, P.; Kaur, R.; Manocha, P.; Prateek; Batra, S.; Singh, P.; Singh, S.; Jindal, P. A Review on Cephalometric Landmark Detection Techniques. Biomed. Signal Process. Control 2021, 66, 102486. [Google Scholar] [CrossRef]
- Yun, H.S.; Hyun, C.M.; Baek, S.H.; Lee, S.-H.; Seo, J.K. Automated 3D Cephalometric Landmark Identification Using Computerized Tomography. arXiv 2020, arXiv:2101.05205. [Google Scholar]
- Silva, T.P.; Hughes, M.M.; Menezes, L.D.; de Melo, M.D.; Takeshita, W.M.; Freitas, P.H. Artificial Intelligence-Based Cephalometric Landmark Annotation and Measurements According to Arnett’s Analysis: Can We Trust a Bot to Do That? Dentomaxillofac. Radiol. 2021, 20200548. [Google Scholar] [CrossRef]
- Yun, H.S.; Jang, T.J.; Lee, S.M.; Lee, S.-H.; Seo, J.K. Learning-Based Local-to-Global Landmark Annotation for Automatic 3D Cephalometry. Phys. Med. Biol. 2020, 65, 085018. [Google Scholar] [CrossRef]
- Kang, S.H.; Jeon, K.; Kim, H.-J.; Seo, J.K.; Lee, S.-H. Automatic Three-Dimensional Cephalometric Annotation System Using Three-Dimensional Convolutional Neural Networks: A Developmental Trial. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2019, 8, 210–218. [Google Scholar] [CrossRef]
- Kute, R.S.; Vyas, V.; Anuse, A. Association of Face and Facial Components Based on CNN and Transfer Subspace Learning for Forensics Applications. SN Comput. Sci. 2020, 1, 1–16. [Google Scholar] [CrossRef]
- Knyaz, V.A.; Maksimov, A.A.; Novikov, M.M.; Urmashova, A.V. Automatic Anthropological Landmarks Recognition and Measurements. ISPAr 2021, 4421, 137–142. [Google Scholar] [CrossRef]
- Guyomarc, P.; Dutailly, B.; Charton, J.; Santos, F.; Desbarats, P. Helene Coqueugniot Anthropological Facial Approximation in Three Dimensions (AFA3D): Computer-Assisted Estimation of the Facial Morphology Using Geometric Morphometrics. J Forensic Sci. 2014, 1502–1516. [Google Scholar] [CrossRef]
- Barbero-García, I.; Pierdicca, R.; Paolanti, M.; Felicetti, A.; Lerma, J.L. Combining Machine Learning and Close-Range Photogrammetry for Infant’s Head 3D Measurement: A Smartphone-Based Solution. Measurement 2021, 182, 109686. [Google Scholar] [CrossRef]
- Wen, Y.; Mingquan, Z.; Pengyue, L.; Guohua, G.; Xiaoning, L.; Kang, L. Ancestry Estimation of Skull in Chinese Population Based on Improved Convolutional Neural Network. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine, BIBM, Online Conference, 16–19 December 2020; pp. 2861–2867. [Google Scholar] [CrossRef]
- Liu, X.; Zou, Y.; Kuang, H.; Ma, X. Face Image Age Estimation Based on Data Augmentation and Lightweight Convolutional Neural Network. Symmetry 2020, 12, 146. [Google Scholar] [CrossRef] [Green Version]
- Fatima, A.; Shahid, A.R.; Raza, B.; Madni, T.M.; Janjua, U.I. State-of-the-Art Traditional to the Machine- and Deep-Learning-Based Skull Stripping Techniques, Models, and Algorithms. J. Digit. Imaging 2020, 33, 1443–1464. [Google Scholar] [CrossRef]
- Tu, P.; Book, R.; Liu, X.; Krahnstoever, N.; Adrian, C.; Williams, P. Automatic Face Recognition from Skeletal Remains. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007. [Google Scholar] [CrossRef]
- Curtner, R.M. Predetermination of the Adult Face. Am. J. Orthod. 1953, 39, 201–217. [Google Scholar] [CrossRef]
- Albert, A.M.; Ricanek, K.; Patterson, E. A Review of the Literature on the Aging Adult Skull and Face: Implications for Forensic Science Research and Applications. Forensic Sci. Int. 2007, 172, 1–9. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, Z.; Wang, X.; Lv, C.; Liu, N. 3D Skull and Face Similarity Measurements Based on a Harmonic Wave Kernel Signature. Vis. Comput. 2020, 37, 749–764. [Google Scholar] [CrossRef]
- Jain, A.; Singh, R.; Vatsa, M. Face Recognition for Forensics. Available online: https://repository.iiitd.edu.in/xmlui/handle/123456789/593?show=fullhttps://repository.iiitd.edu.in/xmlui/handle/123456789/593?show=full (accessed on 1 November 2021).
- Ma, Q.; Kobayashi, E.; Fan, B.; Nakagawa, K.; Sakuma, I.; Masamune, K.; Suenaga, H. Automatic 3D Landmarking Model Using Patch-Based Deep Neural Networks for CT Image of Oral and Maxillofacial Surgery. Int. J. Med Robot. Comput. Assist. Surg. 2020, 16, e2093. [Google Scholar] [CrossRef] [Green Version]
- Tin, M.L. Machine, Discourse and Power: From Machine Learning in Construction of 3D Face to Art and Creativity. Adv. Intell. Syst. Comput. 2020, 517–523. [Google Scholar] [CrossRef]
- Knyaz, V.A.; Kniaz, V.V.; Novikov, M.M.; Galeev, R.M. Machine learning for approximating unknown face—Proquest. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, 43, 857–862. [Google Scholar] [CrossRef]
- Rajesh Kumar, B. Region of interest feature extraction in facial expressions with convolutional neural network classification. Ctact J. Data Sci. Mach. Learn. 2020, 2, 149–152. [Google Scholar]
- Mashouri, P.; Skreta, M.; Phillips, J.; McAllister, D.; Roy, M.; Senkaiahliyan, S.; Brudno, M.; Singh, D. 3D Photography Based Neural Network Craniosynostosis Triaging System. Proc. Mach. Learn. Res. 2020, 136, 226–237. [Google Scholar]
- Lo, L.J.; Yang, C.T.; Ho, C.T.; Liao, C.H.; Lin, H.H. Automatic Assessment of 3-Dimensional Facial Soft Tissue Symmetry Before and After Orthognathic Surgery Using a Machine Learning Model: A Preliminary Experience. Ann. Plast. Surg. 2021, 86, S224–S228. [Google Scholar] [CrossRef]
- Hung, K.; Yeung, A.W.K.; Tanaka, R.; Bornstein, M.M. Current Applications, Opportunities, and Limitations of AI for 3D Imaging in Dental Research and Practice. Int. J. Environ. Res. Public Health 2020, 17, 4424. [Google Scholar] [CrossRef]
- Dalvit Carvalho da Silva, R.; Jenkyn, T.R.; Carranza, V.A. Development of a Convolutional Neural Network Based Skull Segmentation in MRI Using Standard Tesselation Language Models. J. Pers. Med. 2021, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Bekhouche, S.E.; Dornaika, F.; Benlamoudi, A.; Ouafi, A.; Taleb-Ahmed, A. A Comparative Study of Human Facial Age Estimation: Handcrafted Features vs. Deep Features. Multimed. Tools Appl. 2020, 79, 26605–26622. [Google Scholar] [CrossRef]
- Punyani, P.; Gupta, R.; Kumar, A. Neural Networks for Facial Age Estimation: A Survey on Recent Advances. Artif. Intell. Rev. 2019, 53, 3299–3347. [Google Scholar] [CrossRef]
- Liu, H.; Lu, J.; Feng, J.; Zhou, J. Label-Sensitive Deep Metric Learning for Facial Age Estimation. IEEE Trans. Inf. Forensics Secur. 2018, 13, 292–305. [Google Scholar] [CrossRef]
- Lin, H.H.; Lo, L.J.; Chiang, W.C. A Novel Assessment Technique for the Degree of Facial Symmetry before and after Orthognathic Surgery Based on Three-Dimensional Contour Features Using Deep Learning Algorithms. PervasiveHealth Pervasive Comput. Technol. Healthc. 2019, 170–173. [Google Scholar] [CrossRef]
- Makaremi, M.; Lacaule, C.; Mohammad-Djafari, A. Deep Learning and Artificial Intelligence for the Determination of the Cervical Vertebra Maturation Degree from Lateral Radiography. Entropy 2019, 21, 1222. [Google Scholar] [CrossRef] [Green Version]
- Ramanathan, N.; Chellappa, R.; Biswas, S. Computational Methods for Modeling Facial Aging: A Survey. J. Vis. Lang. Comput. 2009, 20, 131–144. [Google Scholar] [CrossRef]
- Dou, P.; Shah, S.K.; Kakadiaris, I.A. End-To-End 3D Face Reconstruction With Deep Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 5908–5917. [Google Scholar]
- Fu, Y.; Guo, G.; Huang, T.S. Age Synthesis and Estimation via Faces: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 1955–1976. [Google Scholar] [CrossRef]
- Sai, P.K.; Wang, J.G.; Teoh, E.K. Facial Age Range Estimation with Extreme Learning Machines. Neurocomputing 2015, 149, 364–372. [Google Scholar] [CrossRef]
- Porto, L.F.; Lima, L.N.; Franco, A.; Pianto, D.; Machado, C.E.; de Barros Vidal, F. Estimating Sex and Age from a Face: A Forensic Approach Using Machine Learning Based on Photo-Anthropometric Indexes of the Brazilian Population. Int. J. Leg. Med. 2020, 134, 2239–2259. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, N.; Chellappa, R.; Biswas, S. Age Progression in Human Faces: A Survey. J. Vis. Lang. Comput. 2009, 15, 3349–3361. [Google Scholar]
- Kaźmierczak, S.; Juszka, Z.; Vandevska-Radunovic, V.; Maal, T.J.; Fudalej, P.; Mańdziuk, J. Prediction of the Facial Growth Direction Is Challenging. Available online: https://arxiv.org/pdf/2110.02316.pdf (accessed on 1 November 2021).
- Mangrulkar, A.; Rane, S.B.; Sunnapwar, V. Automated Skull Damage Detection from Assembled Skull Model Using Computer Vision and Machine Learning. Int. J. Inf. Technol. 2021, 13, 1785–1790. [Google Scholar] [CrossRef]
- Tan, J.S.; Liao, I.Y.; Venkat, I.; Belaton, B.; Jayaprakash, P.T. Computer-Aided Superimposition via Reconstructing and Matching 3D Faces to 3D Skulls for Forensic Craniofacial Identifications. Vis. Comput. 2019, 36, 1739–1753. [Google Scholar] [CrossRef]
- Schmeling, A.; Dettmeyer, R.; Rudolf, E.; Vieth, V.; Geserick, G. Forensic Age Estimation: Methods, Certainty, and the Law. Dtsch. Aerzteblatt Online 2016, 113, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Franklin, D. Forensic Age Estimation in Human Skeletal Remains: Current Concepts and Future Directions. Leg. Med. 2010, 12, 1–7. [Google Scholar] [CrossRef]
- Kotěrová, A.; Navega, D.; Štepanovský, M.; Buk, Z.; Brůžek, J.; Cunha, E. Age Estimation of Adult Human Remains from Hip Bones Using Advanced Methods. Forensic Sci. Int. 2018, 287, 163–175. [Google Scholar] [CrossRef]
- Chia, P.Y.; Coleman, K.K.; Tan, Y.K.; Ong, S.W.X.; Gum, M.; Lau, S.K.; Lim, X.F.; Lim, A.S.; Sutjipto, S.; Lee, P.H.; et al. Detection of Air and Surface Contamination by SARS-CoV-2 in Hospital Rooms of Infected Patients. Nat. Commun. 2020, 11, 2800. [Google Scholar] [CrossRef]
- Singh Sankhla, M.; Kumar, R.; Jadhav, E.B. Artificial Intelligence: Advancing Automation in Forensic Science & Criminal Investigation. J. Seybold Rep. 2020, 15, 2064–2075. [Google Scholar]
- Yadav, J.; Kaur, A. Artificial Neural Network Implementation In Forensic Science. Eur. J. Mol. Clin. Med. 2020, 7, 5935–5939. [Google Scholar]
- Livingston, M. Preventing Racial Bias in Federal AI. JSPG 2020, 16. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting Racial Bias in an Algorithm Used to Manage the Health of Populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarrett, A.; Choo, K.R. The Impact of Automation and Artificial Intelligence on Digital Forensics. WIREs Forensic Sci. 2021. [Google Scholar] [CrossRef]
- Sykes, L.; Bhayat, A.; Bernitz, H. The Effects of the Refugee Crisis on Age Estimation Analysis over the Past 10 Years: A 16-Country Survey. Int. J. Environ. Res. Public Health 2017, 14, 630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.C.; Han, M.; Chi, Y.; Long, H.; Zhang, D.; Yang, J.; Yang, Y.; Chen, T.; Du, S. Accurate Age Classification Using Manual Method and Deep Convolutional Neural Network Based on Orthopantomogram Images. Int. J. Leg. Med. 2021, 135, 1589–1597. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Z.; Dong, X.; Liang, W.; Xue, H.; Zhang, L.; Zhang, Y.; Deng, Z. Forensic Age Estimation for Pelvic X-ray Images Using Deep Learning. Eur. Radiol. 2019, 29, 2322–2329. [Google Scholar] [CrossRef]
- Štepanovský, M.; Ibrová, A.; Buk, Z.; Velemínská, J. Novel Age Estimation Model Based on Development of Permanent Teeth Compared with Classical Approach and Other Modern Data Mining Methods. Forensic Sci. Int. 2017, 279, 72–82. [Google Scholar] [CrossRef]
- Vila-Blanco, N.; Varas-Quintana, P.; Aneiros-Ardao, Á.; Tomás, I.; Carreira, M.J. Automated Description of the Mandible Shape by Deep Learning. Int. J. Comput. Assist. Radiol. Surg. 2021, 1–10. [Google Scholar] [CrossRef]
- De Tobel, J.; Radesh, P.; Vandermeulen, D.; Thevissen, P.W. An Automated Technique to Stage Lower Third Molar Development on Panoramic Radiographs for Age Estimation: A Pilot Study. J. Forensic Odonto-Stomatol. 2017, 35, 42–54. [Google Scholar]
- Merdietio Boedi, R.; Banar, N.; De Tobel, J.; Bertels, J.; Vandermeulen, D.; Thevissen, P.W. Effect of Lower Third Molar Segmentations on Automated Tooth Development Staging Using a Convolutional Neural Network. J. Forensic Sci. 2020, 65, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Štern, D.; Payer, C.; Urschler, M. Automated Age Estimation from MRI Volumes of the Hand. Med. Image Anal. 2019, 58. [Google Scholar] [CrossRef] [PubMed]
- Ortega, R.F.; Irurita, J.; Campo, E.J.E.; Mesejo, P. Analysis of the Performance of Machine Learning and Deep Learning Methods for Sex Estimation of Infant Individuals from the Analysis of 2D Images of the Ilium. Int. J. Leg. Med. 2021, 2659–2666. [Google Scholar] [CrossRef] [PubMed]
- Liew, S.S.; Khalil-Hani, M.; Ahmad Radzi, S.; Bakhteri, R. Gender Classification: A Convolutional Neural Network Approach. Turk. J. Electr. Eng. Comput. Sci. 2016, 24, 1248–1264. [Google Scholar] [CrossRef]
- Kasar, M.M.; Bhattacharyya, D.; Kim, T. Face Recognition Using Neural Network: A Review. Int. J. Secur. Its Appl. 2016, 10, 81–100. [Google Scholar] [CrossRef]
- Pham, C.V.; Lee, S.J.; Kim, S.Y.; Lee, S.; Kim, S.H.; Kim, H.S. Age Estimation Based on 3D Post-Mortem Computed Tomography Images of Mandible and Femur Using Convolutional Neural Networks. PLoS ONE 2021, 16, e0251388. [Google Scholar] [CrossRef]
- Oner, Z.; Turan, M.K.; Oner, S.; Secgin, Y.; Sahin, B. Sex Estimation Using Sternum Part Lenghts by Means of Artificial Neural Networks. Forensic Sci. Int. 2019, 301, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Farhadian, M.; Salemi, F.; Saati, S.; Nafisi, N. Dental Age Estimation Using the Pulp-to-Tooth Ratio in Canines by Neural Networks. Imaging Sci. Dent. 2019, 49, 19–26. [Google Scholar] [CrossRef]
- Etli, Y.; Asirdizer, M.; Hekimoglu, Y.; Keskin, S.; Yavuz, A. Sex Estimation from Sacrum and Coccyx with Discriminant Analyses and Neural Networks in an Equally Distributed Population by Age and Sex. Forensic Sci. Int. 2019, 303, 109955. [Google Scholar] [CrossRef]
- Chen, X.; Lian, C.; Deng, H.H.; Kuang, T.; Lin, H.Y.; Xiao, D.; Gateno, J.; Shen, D.; Xia, J.J.; Yap, P.T. Fast and Accurate Craniomaxillofacial Landmark Detection via 3D Faster R-CNN. IEEE Trans. Med. Imaging 2021. [Google Scholar] [CrossRef] [PubMed]
- Iyer, T.J.; Rahul, K.; Nersisson, R.; Zhuang, Z.; Joseph Raj, A.N.; Refayee, I. Machine Learning-Based Facial Beauty Prediction and Analysis of Frontal Facial Images Using Facial Landmarks and Traditional Image Descriptors. Comput. Intell. Neurosci. 2021, 2021. [Google Scholar] [CrossRef]
- Baccetti, T.; Franchi, L.; McNamara, J.A. The Cervical Vertebral Maturation (CVM) Method for the Assessment of Optimal Treatment Timing in Dentofacial Orthopedics. Semin. Orthod. 2005, 11, 119–129. [Google Scholar] [CrossRef]
- Corradi, F.; Pinchi, V.; Barsanti, I.; Garatti, S. Probabilistic Classification of Age by Third Molar Development: The Use of Soft Evidence. J. Forensic Sci. 2013, 58, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Focardi, M.; Pinchi, V.; de Luca, F.; Norelli, G.-A. Age Estimation for Forensic Purposes in Italy: Ethical Issues. Int. J. Leg. Med. 2014, 128, 515–522. [Google Scholar] [CrossRef]
- Pinchi, V.; Norelli, G.A.; Caputi, F.; Fassina, G.; Pradella, F.; Vincenti, C. Dental Identification by Comparison of Antemortem and Postmortem Dental Radiographs: Influence of Operator Qualifications and Cognitive Bias. Forensic Sci. Int. 2012, 222, 252–255. [Google Scholar] [CrossRef]
- Pinchi, V.; Pradella, F.; Buti, J.; Baldinotti, C.; Focardi, M.; Norelli, G.A. A New Age Estimation Procedure Based on the 3D CBCT Study of the Pulp Cavity and Hard Tissues of the Teeth for Forensic Purposes: A Pilot Study. J. Forensic Leg. Med. 2015, 36, 150–157. [Google Scholar] [CrossRef]
- Pinchi, V.; Norelli, G.-A.; Pradella, F.; Vitale, G.; Rugo, D.; Nieri, M. Comparison of the Applicability of Four Odontological Methods for Age Estimation of the 14 Years Legal Threshold in a Sample of Italian Adolescents. J. Forensic Odonto-Stomatol. 2012, 30, 17. [Google Scholar]
- Pinchi, V.; de Luca, F.; Focardi, M.; Pradella, F.; Vitale, G.; Ricciardi, F.; Norelli, G.A. Combining Dental and Skeletal Evidence in Age Classification: Pilot Study in a Sample of Italian Sub-Adults. Leg. Med. 2016, 20, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Pinchi, V.; de Luca, F.; Ricciardi, F.; Focardi, M.; Piredda, V.; Mazzeo, E.; Norelli, G.A. Skeletal Age Estimation for Forensic Purposes: A Comparison of GP, TW2 and TW3 Methods on an Italian Sample. Forensic Sci. Int. 2014, 238, 83–90. [Google Scholar] [CrossRef]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Using Artificial Intelligence to Detect COVID-19 and Community-Acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology 2020, 296, E65–E71. [Google Scholar] [CrossRef]
- Sessa, F.; Bertozzi, G.; Cipolloni, L.; Baldari, B.; Cantatore, S.; D’Errico, S.; di Mizio, G.; Asmundo, A.; Castorina, S.; Salerno, M.; et al. Clinical-Forensic Autopsy Findings to Defeat COVID-19 Disease: A Literature Review. J. Clin. Med. 2020, 9, 2026. [Google Scholar] [CrossRef]
- Shamout, F.E.; Shen, Y.; Wu, N.; Kaku, A.; Park, J.; Makino, T.; Jastrzębski, S.; Witowski, J.; Wang, D.; Zhang, B.; et al. An Artificial Intelligence System for Predicting the Deterioration of COVID-19 Patients in the Emergency Department. NPJ Digit. Med. 2021, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Esposito, M.; Messina, G.; Di Mizio, G.; Di Nunno, N.; Salerno, M. Sudden Death in Adults: A Practical Flow Chart for Pathologist Guidance. Healthcare 2021, 9, 870. [Google Scholar] [CrossRef]
- Ali, A.R.; Al-Nakib, L.H. The Value of Lateral Cephalometric Image in Sex Identification. J. Baghdad Coll. Dent. 2013, 325, 1–5. [Google Scholar] [CrossRef]
- Patil, K.R.; Mody, R.N. Determination of Sex by Discriminant Function Analysis and Stature by Regression Analysis: A Lateral Cephalometric Study. Forensic Sci. Int. 2005, 147, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Krishan, K. Anthropometry in Forensic Medicine and Forensic Science-Forensic Anthropometry. Internet J. Forensic Sci. 2007, 2, 95–97. [Google Scholar]
- Kim, M.-J.; Liu, Y.; Oh, S.H.; Ahn, H.-W.; Kim, S.-H.; Nelson, G. Automatic Cephalometric Landmark Identification System Based on the Multi-Stage Convolutional Neural Networks with CBCT Combination Images. Sensors 2021, 21, 505. [Google Scholar] [CrossRef]
- Leonardi, R.; lo Giudice, A.; Isola, G.; Spampinato, C. Deep Learning and Computer Vision: Two Promising Pillars, Powering the Future in Orthodontics. Semin. Orthod. 2021, 27, 62–68. [Google Scholar] [CrossRef]
- Bouletreau, P.; Makaremi, M.; Ibrahim, B.; Louvrier, A.; Sigaux, N. Artificial Intelligence: Applications in Orthognathic Surgery. J. Stomatol. Oral Maxillofac. Surg. 2019, 120, 347–354. [Google Scholar] [CrossRef]
- Hung, K.; Montalvao, C.; Tanaka, R.; Kawai, T.; Bornstein, M.M. The Use and Performance of Artificial Intelligence Applications in Dental and Maxillofacial Radiology: A Systematic Review. Dentomaxillofac. Radiol. 2019, 49. [Google Scholar] [CrossRef] [PubMed]
- Schwendicke, F.; Chaurasia, A.; Arsiwala, L.; Lee, J.-H.; Elhennawy, K.; Jost-Brinkmann, P.-G.; Demarco, F.; Krois, J. Deep Learning for Cephalometric Landmark Detection: Systematic Review and Meta-Analysis. Clin. Oral Investig. 2021, 25, 4299–4309. [Google Scholar] [CrossRef]
- Koberová, K.; Thurzo, A.; Dianišková, S. Evaluation of Prevalence of Facial Asymmetry in Population According to the Analysis of 3D Face-Scans. Lek. Obz. 2015, 64, 101–104. [Google Scholar]
- Tanikawa, C.; Yamashiro, T. Development of Novel Artificial Intelligence Systems to Predict Facial Morphology after Orthognathic Surgery and Orthodontic Treatment in Japanese Patients. Sci. Rep. 2021, 11, 15853. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Choi, K.; Jung, H.; Duan, L. Deep Learning for Facial Beauty Prediction. Information 2020, 11, 391. [Google Scholar] [CrossRef]
- Dang, H.; Liu, F.; Stehouwer, J.; Liu, X.; Jain, A.K. On the Detection of Digital Face Manipulation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 5781–5790. [Google Scholar]
- Kittler, J.; Koppen, P.; Kopp, P.; Huber, P.; Ratsch, M. Conformal Mapping of a 3d Face Representation onto a 2D Image for CNN Based Face Recognition. In Proceedings of the 2018 International Conference on Biometrics (ICB), Gold Coast, Australia, 20–23 February 2018; pp. 124–131. [Google Scholar] [CrossRef]
- Turchetta, B.J.; Fishman, L.S.; Subtelny, J.D. Facial Growth Prediction: A Comparison of Methodologies. Am. J. Orthod. Dentofac. Orthop. 2007, 132, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Hirschfeld, W.J.; Moyers, R.E. Prediction of Craniofacial Growth: The State of the Art. Am. J. Orthod. 1971, 60, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Kaźmierczak, S.; Juszka, Z.; Fudalej, P.; Mańdziuk, J. Prediction of the Facial Growth Direction with Machine Learning Methods. arXiv 2021, arXiv:2106.10464. [Google Scholar]
- Yang, C.; Rangarajan, A.; Ranka, S. Visual Explanations From Deep 3D Convolutional Neural Networks for Alzheimer’s Disease Classification. AMIA Annu. Symp. Proc. 2018, 2018, 1571. [Google Scholar] [PubMed]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR, Honolulu, HI, USA, 21–26 July 2016; pp. 2261–2269. [Google Scholar]
- de Jong, M.A.; Gül, A.; de Gijt, J.P.; Koudstaal, M.J.; Kayser, M.; Wolvius, E.B.; Böhringer, S. Automated Human Skull Landmarking with 2D Gabor Wavelets. Phys. Med. Biol. 2018, 63. [Google Scholar] [CrossRef]
- Arık, S.; Ibragimov, B.; Xing, L. Fully Automated Quantitative Cephalometry Using Convolutional Neural Networks. J. Med Imaging 2017, 4, 014501. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Deng, H.; Lian, C.; Chen, X.; Xiao, D.; Ma, L.; Chen, X.; Kuang, T.; Gateno, J.; Yap, P.-T.; et al. SkullEngine: A Multi-Stage CNN Framework for Collaborative CBCT Image Segmentation and Landmark Detection. In International Workshop on Machine Learning in Medical Imaging; Springer: Cham, The Netherlands, 2021; pp. 606–614. [Google Scholar] [CrossRef]
- Sessa, F.; Salerno, M.; Pomara, C.; Sessa, F.; Salerno, M.; Pomara, C. Autopsy Tool in Unknown Diseases: The Experience with Coronaviruses (SARS-CoV, MERS-CoV, SARS-CoV-2). Medicina 2021, 57, 309. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Minnema, J.; Batenburg, K.J.; Forouzanfar, T.; Hu, F.J.; Wu, G. Multiclass CBCT Image Segmentation for Orthodontics with Deep Learning. J. Dent. Res. 2021, 100, 943–949. [Google Scholar] [CrossRef]
- Jang, T.J.; Kim, K.C.; Cho, H.C.; Seo, J.K. A Fully Automated Method for 3D Individual Tooth Identification and Segmentation in Dental CBCT. IEEE Trans. Pattern Anal. Mach. Intell. 2021. [Google Scholar] [CrossRef]
- Qiu, B.; van der Wel, H.; Kraeima, J.; Glas, H.H.; Guo, J.; Borra, R.J.H.; Witjes, M.J.H.; van Ooijen, P.M.A. Robust and Accurate Mandible Segmentation on Dental CBCT Scans Affected by Metal Artifacts Using a Prior Shape Model. J. Pers. Med. 2021, 11, 364. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; van der Wel, H.; Kraeima, J.; Glas, H.H.; Guo, J.; Borra, R.J.H.; Witjes, M.J.H.; van Ooijen, P.M.A. Mandible Segmentation of Dental CBCT Scans Affected by Metal Artifacts Using Coarse-to-Fine Learning Model. J. Pers. Med. 2021, 11, 560. [Google Scholar] [CrossRef] [PubMed]
- Funatsu, M.; Sato, K.; Mitani, H. Effects of Growth Hormone on Craniofacial GrowthDuration of Replacement Therapy. Angle Orthod. 2006, 76, 970–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantu, G.; Buschang, P.H.; Gonzalez, J.L. Differential Growth and Maturation in Idiopathic Growth-Hormone-Deficient Children. Eur. J. Orthod. 1997, 19, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Konfino, R.; Pertzelan, A.; Laron, Z. Cephalometric Measurements of Familial Dwarfism and High Plasma Immunoreactive Growth Hormone. Am. J. Orthod. 1975, 68, 196–201. [Google Scholar] [CrossRef]
- Grippaudo, C.; Paolantonio, E.G.; Antonini, G.; Saulle, R.; la Torre, G.; Deli, R. Association between Oral Habits, Mouth breathing and Malocclusion. Acta Otorhinolaryngol. Italy 2016, 36, 386. [Google Scholar] [CrossRef]
- Ranggang, B.M.; Armedina, R.N. CrossMark Comparison of Parents Knowledge of Bad Habits and the Severity Maloclusion of Children in Schools with Different Social Levels. J. Dentomaxillofacial Sci. 2020, 5, 48–51. [Google Scholar] [CrossRef]
- Arcari, S.R.; Biagi, R. Oral Habits and Induced Occlusal-Skeletal Disarmonies. J. Plast. Dermatol. 2015, 11, 1. [Google Scholar]
- Hammond, P.; Hannes, F.; Suttie, M.; Devriendt, K.; Vermeesch, J.R.; Faravelli, F.; Forzano, F.; Parekh, S.; Williams, S.; McMullan, D.; et al. Fine-Grained Facial Phenotype–Genotype Analysis in Wolf–Hirschhorn Syndrome. Eur. J. Hum. Genet. 2011, 20, 33–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartsfield, J.K. The Importance of Analyzing Specific Genetic Factors in Facial Growth for Diagnosis and Treatment Planning. Available online: https://www.researchgate.net/profile/James-Hartsfield/publication/235916088_The_Importance_of_Analyzing_Specific_Genetic_Factors_in_Facial_Growth_for_Diagnosis_and_Treatment_Planning/links/00b7d5141dd8fd2d82000000/The-Importance-of-Analyzing-Specific-Genetic-Factors-in-Facial-Growth-for-Diagnosis-and-Treatment-Planning.pdf (accessed on 1 November 2021).
- Hartsfield, J.K.; Morford, L.A.; Otero, L.M.; Fardo, D.W. Genetics and Non-Syndromic Facial Growth. J. Pediatr. Genet. 2015, 2, 9–20. [Google Scholar] [CrossRef] [Green Version]










| Parameter | Setting |
|---|---|
| Sensor Type | Amorphous Silicon Flat Panel Sensor with Csl Scintillator |
| Grayscale Resolution | 16-bit |
| Voxel Size | 0.3 mm, |
| Collimation | Electronically controlled fully adjustable collimation |
| Scan Time | 17.8 s |
| Exposure Type | Pulsed |
| Field-of-View | 23 cm × 17 cm |
| Reconstruction Shape | Cylinder |
| Reconstruction Time | Less than 30 s |
| Output | DICOM |
| Patient Position | Seated |
| Area of Forensic Research | Proposed Method | Metrics |
|---|---|---|
| Biological age determination | Regression model by 3D deep CNN | MAE, MSE |
| Sex determination | Deep 3D CNN—conv.layers and outputs class probabilities for both targets | CM such as precision, recall and F1 score |
| 3D cephalometric analysis | Object detection model on 3D CNN that auto.estimates cephalom.measurements | MAE, MSE |
| Face prediction from skull | model on Generative Adversarial Network that synthesize soft/hard tissues | slice-wise Frechet Inception Distance |
| Facial growth prediction | Based on methods stated above 1 | another 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thurzo, A.; Kosnáčová, H.S.; Kurilová, V.; Kosmeľ, S.; Beňuš, R.; Moravanský, N.; Kováč, P.; Kuracinová, K.M.; Palkovič, M.; Varga, I. Use of Advanced Artificial Intelligence in Forensic Medicine, Forensic Anthropology and Clinical Anatomy. Healthcare 2021, 9, 1545. https://0-doi-org.brum.beds.ac.uk/10.3390/healthcare9111545
Thurzo A, Kosnáčová HS, Kurilová V, Kosmeľ S, Beňuš R, Moravanský N, Kováč P, Kuracinová KM, Palkovič M, Varga I. Use of Advanced Artificial Intelligence in Forensic Medicine, Forensic Anthropology and Clinical Anatomy. Healthcare. 2021; 9(11):1545. https://0-doi-org.brum.beds.ac.uk/10.3390/healthcare9111545
Chicago/Turabian StyleThurzo, Andrej, Helena Svobodová Kosnáčová, Veronika Kurilová, Silvester Kosmeľ, Radoslav Beňuš, Norbert Moravanský, Peter Kováč, Kristína Mikuš Kuracinová, Michal Palkovič, and Ivan Varga. 2021. "Use of Advanced Artificial Intelligence in Forensic Medicine, Forensic Anthropology and Clinical Anatomy" Healthcare 9, no. 11: 1545. https://0-doi-org.brum.beds.ac.uk/10.3390/healthcare9111545