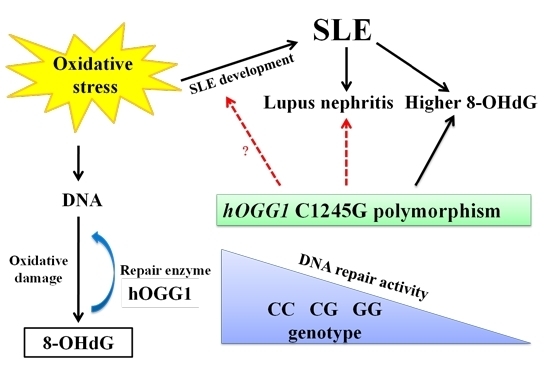
The Role of hOGG1 C1245G Polymorphism in the Susceptibility to Lupus Nephritis and Modulation of the Plasma 8-OHdG in Patients with Systemic Lupus Erythematosus †
Abstract
:1. Introduction
2. Results and Discussion
2.1. Distributions of hOGG1 C1245G Polymorphism in Healthy Controls and SLE Patients
2.2. Levels of Plasma 8-OHdG in Healthy Controls and SLE Patients Based on the hOGG1 Gene Polymorphisms

| Subjects (Case Number, %) | hOGG1 C1245G Polymorphisms | p-Value | |
|---|---|---|---|
| hOGG1 1245 CC or CG Genotype | hOGG1 GG Genotype | ||
| Healthy controls (n = 45, 100) | 21 (46.7)—Group A | 24 (53.3)—Group B | 0.017 * |
| SLE patients (n = 85, 100) | 58 (68.2)—Group C | 27 (31.8)—Group D | |
| Plasma 8-OhdG (M ± SD, ng/mL) | Plasma 8-OHdG (M ± SD, ng/mL) | p-Value | ||
|---|---|---|---|---|
| Healthy controls (n = 45) | 0.157 ± 0.038 | SLE patients (n = 85) | 0.225 ± 0.082 | <0.001 [20] |
| hOGG1 C1245G polymorphisms | hOGG1 C1245G polymorphisms | |||
| Group A (CC or CG genotypes, n = 21) | 0.161 ± 0.036 | Group C (CC or CG genotypes, n = 58) | 0.217 ± 0.059 | <0.001 * |
| Group B (GG genotype, n = 24) | 0.155 ± 0.041 | Group D (GG genotype, n = 27) | 0.243 ± 0.117 | 0.001 * |
| p-value | 0.614 ** | p-value | 0.289 ** | <0.001 *** |

2.3. Distributions of hOGG1 C1245G Polymorphisms in SLE Patients with or without Lupus Nephritis
| SLE Patients | hOGG1 Gene Polymorphisms (Case Number, %) | p-Value | |
|---|---|---|---|
| hOGG1 1245 CC or CG Genotype (n = 58, 100%) | hOGG1 GG Genotype (n = 27, 100%) | ||
| Without nephritis | 40 (69.0%)—Group I | 12 (44.4%)—Group II | 0.031 * |
| With nephritis | 18 (31.0%)—Group III | 15 (55.6%)—Group IV | |
2.4. Plasma Levels of 8-OHdG in SLE Patients as Related to the Clinical Presentations of Lupus Nephritis and hOGG1 C1245G Polymorphism
| Plasma 8-OhdG (M ± SD, ng/mL) | Plasma 8-OhdG (M ± SD, ng/mL) | p-Value | ||
|---|---|---|---|---|
| SLE patients without nephritis (n = 52) | 0.218 ± 0.060 | SLE patients with nephritis (n = 33) | 0.237 ± 0.109 | 0.313 |
| Group I hOGG1 1245 CC or CG genotype (n = 40) | 0.224 ± 0.064 | Group III hOGG1 1245 CC or CG genotype (n = 18) | 0.201 ± 0.046 | 0.165 * |
| Group II GG genotype (Cys/Cys hOGG1) (n = 12) | 0.197 ± 0.040 | Group IV GG genotype (Cys/Cys hOGG1) (n = 15) | 0.280 ± 0.145 | 0.050 * |
| p-value | 0.164 ** | 0.037 ** | 0.054 *** | |
| 0.020 **** |

3. Experimental Section
3.1. Recruitment of SLE Patients and Healthy Controls
3.2. Blood Sample Collection, Plasma Separation and Leukocyte DNA Preparation
3.3. Determination of 8-OHdG in Plasma by ELISA
3.4. Analysis of hOGG1 C1245G Polymorphisms
3.5 Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Perl, A. Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity 2010, 43, 1–6. [Google Scholar] [PubMed]
- Li, K.J.; Wu, C.H.; Hsieh, S.C.; Lu, M.C.; Tsai, C.Y.; Yu, C.L. Deranged bioenergetics and defective redox capacity in T lymphocytes and neutrophils are related to cellular dysfunction and increased oxidative stress in patients with active systemic lupus erythematosus. Clin. Dev. Immunol. 2012, 2012. [Google Scholar] [CrossRef]
- Shah, D.; Wanchu, A.; Bhatnagar, A. Interaction between oxidative stress and chemokines: Possible pathogenic role in systemic lupus erythematosus and rheumatoid arthritis. Immunobiology 2011, 216, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Pierangeli, S.S.; Papalardo, E.; Ansari, G.A.; Khan, M.F. Markers of oxidative and nitrosative stress in systemic lupus erythematosus: Correlation with disease activity. Arthritis Rheumatol. 2010, 62, 2064–2072. [Google Scholar]
- Domann, F.E. Aberrant free radical biology is a unifying theme in the etiology and pathogenesis of major human diseases. Int. J. Mol. Sci. 2013, 14, 8491–8495. [Google Scholar] [CrossRef] [PubMed]
- Neeley, W.L.; Essigmann, J.M. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006, 19, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar] [PubMed]
- Boiteux, S.; Radicella, J.P. The human OGG1 gene: Structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 2000, 377, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Shinmura, K.; Tosaka, M.; Tani, M.; Kim, S.R.; Sugimura, H.; Nohmi, T.; Kasai, H.; Yokota, J. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene 1998, 16, 3219–3225. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.K.; Hsieh, W.A.; Tsai, M.H.; Chen, C.C.; Hong, A.I.; Wei, Y.H.; Chang, W.P. Age-associated decrease of oxidative repair enzymes, human 8-oxoguanine DNA glycosylases (hOGG1), in human aging. J. Radiat. Res. 2003, 44, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, X.; Zhang, H.; Guo, W.; Cai, Z.; Chen, H.; Zhang, K.; Zhu, D.; Wang, Y. Functional polymorphism of hOGG1 gene is associated with type 2 diabetes mellitus in Chinese population. Mol. Cell. Endocrinol. 2010, 325, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Coppede, F.; Migheli, F.; Ceravolo, R.; Bregant, E.; Rocchi, A.; Petrozzi, L.; Unti, E.; Lonigro, R.; Siciliano, G.; Migliore, L. The hOGG1 Ser326Cys polymorphism and Huntington’s disease. Toxicology 2010, 278, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.F.; Xu, Y.J.; Xie, J.G.; Zhang, Z.X. hOGG1 Ser326Cys and XRCC1 Arg399Gln polymorphisms associated with chronic obstructive pulmonary disease. Chin. Med. J. 2009, 122, 960–966. [Google Scholar] [PubMed]
- Tanrikulu, S.; Dogru-Abbasoglu, S.; Ozderya, A.; Ademoglu, E.; Karadag, B.; Erbil, Y.; Uysal, M. The 8-oxoguanine DNA N-glycosylase 1 (hOGG1) Ser326Cys variant affects the susceptibility to Graves’ disease. Cell Biochem. Funct. 2011, 29, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Chen, L.; Tockman, M.S.; Elahi, A.; Lazarus, P. The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA repair enzyme and its association with lung cancer risk. Pharmacogenetics 2004, 14, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Wang, L.S.; Chou, T.Y.; Hsu, W.H.; Lin, H.C.; Lee, S.Y.; Lee, M.H.; Chang, S.C.; Wei, Y.H. Cigarette smoking and hOGG1 Ser326Cys polymorphism are associated with 8-OHdG accumulation on mitochondrial DNA in thoracic esophageal squamous cell carcinoma. Ann. Surg. Oncol. 2013, 20, S379–S388. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.Z.; Gheita, T.A.; Kenawy, S.A.; Fahim, A.T.; El-Sorougy, I.M.; Abdou, M.S. Oxidative stress in systemic lupus erythematosus and rheumatoid arthritis patients: Relationship to disease manifestations and activity. Int. J. Rheum. Dis. 2011, 14, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Chen, H.; Tao, J.; Guo, W.; Liu, X.; Zheng, B.; Sun, W.; Wang, Y. Association of base excision repair gene polymorphisms with ESRD risk in a Chinese population. Oxid. Med. Cell. Longev. 2012, 2012, 928421. [Google Scholar] [PubMed]
- Tarng, D.C.; Tsai, T.J.; Chen, W.T.; Liu, T.Y.; Wei, Y.H. Effect of human OGG1 1245C→G gene polymorphism on 8-hydroxy-2'-deoxyguanosine levels of leukocyte DNA among patients undergoing chronic hemodialysis. J. Am. Soc. Nephrol. 2001, 12, 2338–2347. [Google Scholar] [PubMed]
- Lee, H.T.; Lin, C.S.; Lee, C.S.; Tsai, C.Y.; Wei, Y.H. Increased 8-hydroxy-2'-deoxyguanosine in plasma and decreased mRNA expression of human 8-oxoguanine DNA glycosylase 1, antioxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes in leukocytes in patients with systemic lupus erythematosus. Clin. Exp. Immunol. 2014, 176, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, H.; Kohno, T.; Wakai, K.; Nagura, K.; Genka, K.; Igarashi, H.; Morris, B.J.; Baba, S.; Ohno, Y.; Gao, C.; et al. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol. Biomark. Prev. 1999, 8, 669–674. [Google Scholar]
- Wang, C.L.; Lin, T.H.; Lin, H.Y.; Sheu, S.H.; Yu, M.L.; Hsiao, P.J.; Lin, K.D.; Hsu, C.; Yang, Y.H.; Shin, S.J. The 8-oxoguanine glycosylase 1 (hOGG1) Ser326Cys variant affects the susceptibility to multi-vessel disease in Taiwan coronary artery disease patients. Thromb. Res. 2010, 126, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Sasaki, T.; Arakawa, S.; Fujimoto, S.; Horike, H.; Hatta, H.; Kashihara, N. hOGG1 polymorphism correlates with progression of IgA nephropathy. Nephrology 2001, 6, A10–A11. [Google Scholar] [CrossRef]
- Hochberg, M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheumatol. 1997, 40, 1725. [Google Scholar] [CrossRef]
- Lee, H.T.; Lin, C.S.; Chen, W.S.; Liao, H.T.; Tsai, C.Y.; Wei, Y.H. Leukocyte mitochondrial DNA alteration in systemic lupus erythematosus and its relevance to the susceptibility to lupus nephritis. Int. J. Mol. Sci. 2012, 13, 8853–8868. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.S.; Kuo, C.L.; Cheng, W.L.; Huang, C.S.; Lee, C.F.; Wei, Y.H. Alteration of the copy number of mitochondrial DNA in leukocytes of patients with hyperlipidemia. Ann. N. Y. Acad. Sci. 2005, 1042, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Takeda, H.; Otake, S.; Yokozawa, J.; Nishise, S.; Fujishima, S.; Orii, T.; Fukui, T.; Takano, J.; Sasaki, Y.; et al. Increased plasma levels of 8-hydroxydeoxyguanosine are associated with development of colorectal tumors. J. Clin. Biochem. Nutr. 2010, 47, 59–63. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-T.; Lin, C.-S.; Lee, C.-S.; Tsai, C.-Y.; Wei, Y.-H. The Role of hOGG1 C1245G Polymorphism in the Susceptibility to Lupus Nephritis and Modulation of the Plasma 8-OHdG in Patients with Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2015, 16, 3757-3768. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16023757
Lee H-T, Lin C-S, Lee C-S, Tsai C-Y, Wei Y-H. The Role of hOGG1 C1245G Polymorphism in the Susceptibility to Lupus Nephritis and Modulation of the Plasma 8-OHdG in Patients with Systemic Lupus Erythematosus. International Journal of Molecular Sciences. 2015; 16(2):3757-3768. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16023757
Chicago/Turabian StyleLee, Hui-Ting, Chen-Sung Lin, Chyou-Shen Lee, Chang-Youh Tsai, and Yau-Huei Wei. 2015. "The Role of hOGG1 C1245G Polymorphism in the Susceptibility to Lupus Nephritis and Modulation of the Plasma 8-OHdG in Patients with Systemic Lupus Erythematosus" International Journal of Molecular Sciences 16, no. 2: 3757-3768. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16023757