Genetically Encoded Voltage Indicators in Circulation Research
Abstract
:1. Measuring Membrane Potentials—Principles and Properties
2. Approaches of Genetically Encoded Voltage Indicators
2.1. GEVI (Genetically Encoded Voltage Indicators) Based on Voltage Sensitive Conformational Changes

| # | GEVI | Voltage Sensing Principle | FRET Pair (1–3, 8) Fluorescent Protein (4–7) | Principle Design and Operation with Permission from Elsevier [29] | ∆R/R per 100 mV (1–3) ∆F/F per 100 mV (4–8) | Detection Range | Temporal Response (on); Jump from –70 mV to at least +30 mV | Application in Circulation Research/Comments |
|---|---|---|---|---|---|---|---|---|
| 1 | VSFP2.3 [30] | conformational change by phosphatase of sea squirt (Ciona intestinalis) [22] | mCerulian (CFP) and Citrine (YFP) [30] |  | 13.3% ± 3.4% [26], 10% ± 1% [27] | half activation ~–40 mV [27] | biexponential 2.5 ± 0.5 and 25 ± 3 ms −70 to +60 mV and 35 °C [26] | optical mapping in transgenic mouse heart [31] |
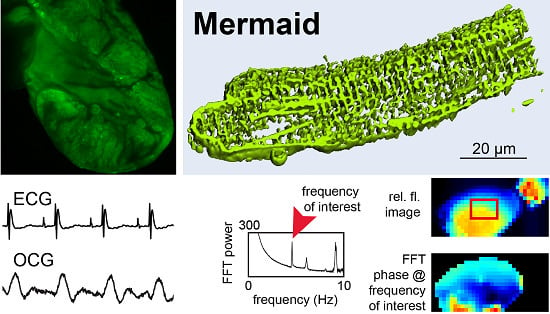
| 2 | Mermaid [25] | Umi Kinoko (mUKG) and the monomeric Kusabira Orange (mKOκ) [25] | 12.9 ± 4.8% [26] | half activation ~–40 mV [25] | biexponential 2.5 ± 0.5 ms (23% ± 5%) and 25 ± 3 ms at 35 °C [26] | cardiotoxicity screens in vivo (zebrafish) [32] and in isolated cardiac myocytes (rat) [33]; optical mapping in transgenic mouse heart (this paper) | ||
| 3 | VSFP-CR [27] | Clover and mRuby2 [27] | 13 ± 1% [27] | half activation ~–40 mV [27] | biexponential 5.4 ± 0.8 and 59.5 ± 5.5 ms at 20 °C [27] | measurements in stem cell derived cardiomyocytes for phenotyping (this paper) | ||
| 4 | ArcLight [34] | super ecliptic pH luorin (A227D) GFP [35] |  | ~32% [36] | half activation ~–25 mV [34] | biexponential ~17.4 ms (~39%) and ~123 ms at 23 °C [36] | stem cell derived cardiomyocyte phenotyping [37] | |
| 5 | ASAP1 [38] | chicken (Gallus gallus) voltage-sensitive phosphatase [38] | circularly permutated GFP |  | ~29% [39] | –120 to −50 mV superlinear and –50 to 50 mV linear response [38] | biexponential 2.1 ± 0.2 ms (60.2%± 1.2%) and 71.5 ± 1.6 ms [38] | to be done (t.b.d.) |
| 6 | Arch (D95N) [40] | microbial rhodopsin proton pumps | modified Archae-rhodopsin 3 [40] |  | ~40% [36] | –150 to +150 mV almost linear response [40] | biexponential < 0.5 ms (~20%) and ~41 ms [40] | mapping of membrane potential in transgenic zebrafish heart [41] |
| 7 | QuasAr2 [36] | modified Archae-rhodopsin 3 [36] | 90% ± 2% [36] | –100 to +50 mV almost linear response [36] | biexponential 1.2 ± 0.1 ms (68%) and 11.8 ± 1.5 ms; similar on rising and falling edge [36] | t.b.d./most bathochromic GEVI (exitation 590 nm, emission 715 nm); although high sensitivity, fluorescence intensity is ∼50-fold dimer than GFP [39] | ||
| 8 | MacQ [42] | voltage induced shifts in the absorption spectrum of Leptosphaeria maculans rhodopsin results in quenching of the attached mCitrine or mOrange2 [42], although FRET is happening, only the intensity change of the donor is measured |  | ~20% [42] | –100 to 0 mV almost linear response, leveling out at 0 mV [42] | biexponential 2.8 ± 0.2 ms (74% ± 2%) and 71 ± 3 ms (26% ± 2%) for mCitrine and 2.9 ± 0.1 ms (96% ± 1%) and 115 ± 10 ms (4% ± 1%) for mOrange2 [42] | t.b.d./based on the same principle a palette of multicolored GEVI have been introduced [39] | |
2.2. Microbial Opsin-Based GEVIs
2.3. Sensing Non-Linear Optical Properties of Fluorescent Proteins
3. Examples of GEVIs in Circulation Research
3.1. Cardiotoxicity Screens
3.2. Stem Cell Derived Cardiomyocyte Phenotyping
3.3. Optical Mapping in Transgenic Heart
4. Summary and Perspective

Acknowledgments
Conflicts of Interest
References
- Matteucci, C. Sur un phenomene physiologique produit par les muscles en contraction. Ann. Chim. Phys. 1842, 6, 339–341. (In French) [Google Scholar]
- Du Bois-Reymond, E. Untersuchungen über Thierische Elektricitat; Verlag von G. Reimer: Berlin, Germany, 1848. (In German) [Google Scholar]
- Von Helmholtz, H. Handbuch der physiologischen Optik; Leopold Voss: Leipzig, Germany, 1867. (In German) [Google Scholar]
- Cole, K.S. Mostly membranes. Annu. Rev. Physiol. 1979, 41, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hamill, O.P.; Marty, A.; Neher, E.; Sakmann, B.; Sigworth, F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 1981, 391, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Sigworth, F.J. The patch clamp is more useful than anyone had expected. Fed. Proc. 1986, 45, 2673–2677. [Google Scholar] [PubMed]
- Kaestner, L.; Lipp, P. Towards imaging the dynamics of protein signalling. In Imaging Cellular and Molecular Biological Functions; Shorte, S.L., Frischknecht, F., Eds.; Springer: Heidelberg, Germany, 2007; pp. 289–312. [Google Scholar]
- Stepnoski, R.A.; LaPorta, A.; Raccuia-Behling, F.; Blonder, G.E.; Slusher, R.E.; Kleinfeld, D. Noninvasive detection of changes in membrane potential in cultured neurons by light scattering. Proc. Natl. Acad. Sci. USA 1991, 88, 9382–9386. [Google Scholar] [CrossRef] [PubMed]
- Hinner, M.J.; Hübener, G.; Fromherz, P. Enzyme-induced staining of biomembranes with voltage-sensitive fluorescent dyes. J. Phys. Chem. 2004, 108, 2445–2453. [Google Scholar] [CrossRef] [PubMed]
- Hinner, M.J.; Hübener, G.; Fromherz, P. Genetic targeting of individual cells with a voltage-sensitive dye through enzymatic activation of membrane binding. ChemBioChem 2006, 7, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Chanda, B.; Blunck, R.; Faria, L.C.; Schweizer, F.E.; Mody, I.; Bezanilla, F. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat. Neurosci. 2005, 8, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- DiFranco, M.; Capote, J.; Quinonez, M.; Vergara, J.L. Voltage-dependent dynamic FRET signals from the transverse tubules in mammalian skeletal muscle fibers. J. Gen. Physiol. 2007, 130, 581–600. [Google Scholar] [CrossRef] [PubMed]
- Ghitani, N.; Bayguinov, P.O.; Ma, Y.; Jackson, M.B. Single-trial imaging of spikes and synaptic potentials in single neurons in brain slices with genetically encoded hybrid voltage sensor. J. Neurophysiol. 2015, 113, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Sjulson, L.; Miesenböck, G. Rational optimization and imaging in vivo of a genetically encoded optical voltage reporter. J. Neurosci. 2008, 28, 5582–5593. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.H.; Venkatachalam, V.; Cohen, A.E. Temporal dynamics of microbial rhodopsin fluorescence reports absolute membrane voltage. Biophys. J. 2014, 106, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Siegel, M.S.; Isacoff, E.Y. A genetically encoded optical probe of membrane voltage. Neuron 1997, 19, 735–741. [Google Scholar] [CrossRef]
- Guerrero, G.; Siegel, M.S.; Roska, B.; Loots, E.; Isacoff, E.Y. Tuning FlaSh: Redesign of the dynamics, voltage range, and color of the genetically encoded optical sensor of membrane potential. Biophys. J. 2002, 83, 3607–3618. [Google Scholar] [CrossRef]
- Sakai, R.; Repunte-Canonigo, V.; Raj, C.D.; Knöpfel, T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur. J. Neurosci. 2001, 13, 2314–2318. [Google Scholar] [CrossRef] [PubMed]
- Knöpfel, T.; Tomita, K.; Shimazaki, R.; Sakai, R. Optical recordings of membrane potential using genetically targeted voltage-sensitive fluorescent proteins. Methods 2003, 30, 42–48. [Google Scholar] [CrossRef]
- Ataka, K.; Pieribone, V.A. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys. J. 2002, 82, 509–516. [Google Scholar] [CrossRef]
- Baker, B.J.; Lee, H.Y.; Pieribone, V.A.; Cohen, L.B.; Isacoff, E.Y.; Knöpfel, T.; Kosmidis, E.K. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J. Neurosci. Methods 2007, 161, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Iwasaki, H.; Sasaki, M.; Inaba, K.; Okamura, Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 2005, 435, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, I.S.; Moran, M.M.; Chong, J.A.; Clapham, D.E. A voltage-gated proton-selective channel lacking the pore domain. Nature 2006, 440, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, D.; He, Y.; Mutoh, H.; Baker, B.J.; Cohen, L.B.; Akemann, W.; Knöpfel, T. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS ONE 2007, 2, e440. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, H.; Karasawa, S.; Okamura, Y.; Miyawaki, A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat. Methods 2008, 5, 683–685. [Google Scholar] [CrossRef] [PubMed]
- Mutoh, H.; Perron, A.; Dimitrov, D.; Iwamoto, Y.; Akemann, W.; Chudakov, D.M.; Knöpfel, T. Spectrally-resolved response properties of the three most advanced FRET based fluorescent protein voltage probes. PLoS ONE 2009, 4, e4555. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.J.; St-Pierre, F.; Gong, Y.; Marshall, J.D.; Cranfill, P.J.; Baird, M.A.; McKeown, M.R.; Wiedenmann, J.; Davidson, M.W.; Schnitzer, M.J.; et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 2012, 9, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Dorn, T.; Goedel, A.; Lam, J.T.; Haas, J.; Tian, Q.; Herrmann, F.; Bundschu, K.; Dobreva, G.; Schiemann, M.; Dirschinger, R.; et al. Direct Nkx2-5 transcriptional repression of Isl1 controls cardiomyocyte subtype identity. Stem Cells 2015, 33, 1113–1129. [Google Scholar] [CrossRef] [PubMed]
- Knöpfel, T.; Gallero-Salas, Y.; Song, C. Genetically encoded voltage indicators for large scale cortical imaging come of age. Curr. Opin. Chem. Biol. 2015, 27, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Mutoh, H.; Dimitrov, D.; Akemann, W.; Knöpfel, T. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS ONE 2008, 3, e2514. [Google Scholar] [CrossRef] [PubMed]
- Chang Liao, M.-L.; de Boer, T.; Mutoh, H.; Raad, N.S.; Richter, C.; Wagner, E.; Downie, B.R.; Unsöld, B.; Arooj, I.; Streckfuss-Bömeke, K.; et al. Sensing Cardiac Electrical Activity With a Cardiac Myocyte Targeted Optogenetic Voltage Indicator. Circ. Res. 2015, 117, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, H.; Higashijima, S.I.; Miyawaki, A.; Okamura, Y. Visualizing voltage dynamics in zebrafish heart. J. Physiol. 2010, 588, 2017–2021. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Oberhofer, M.; Ruppenthal, S.; Scholz, A.; Buschmann, V.; Tsutsui, H.; Miyawaki, A.; Zeug, A.; Lipp, P.; Kaestner, L. Optical action potential screening on adult ventricular myocytes as an alternative QT-screen. Cell Physiol. Biochem. 2011, 27, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Han, Z.; Platisa, J.; Wooltorton, J.R.A.; Cohen, L.B.; Pieribone, V.A. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 2012, 75, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Miesenbock, G.; de Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Hochbaum, D.R.; Zhao, Y.; Farhi, S.L.; Klapoetke, N.; Werley, C.A.; Kapoor, V.; Zou, P.; Kralj, J.M.; Maclaurin, D.; Smedemark-Margulies, N.; et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 2014, 11, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Leyton-Mange, J.S.; Mills, R.W.; Macri, V.S.; Jang, M.Y.; Butte, F.N.; Ellinor, P.T.; Milan, D.J. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep. 2014, 2, 163–170. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, F.; Marshall, J.D.; Yang, Y.; Gong, Y.; Schnitzer, M.J.; Lin, M.Z. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014, 17, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Zhao, Y.; Douglass, A.D.; Hochbaum, D.R.; Brinks, D.; Werley, C.A.; Harrison, D.J.; Campbell, R.E.; Cohen, A.E. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kralj, J.M.; Douglass, A.D.; Hochbaum, D.R.; Maclaurin, D.; Cohen, A.E. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods 2012, 9, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.H.; Kralj, J.M.; Douglass, A.D.; Engert, F.; Cohen, A.E. Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific developmental transitions in ionic currents. Front. Physiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Wagner, M.J.; Li, J.Z.; Schnitzer, M.J. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.G.; Perron, A.; Mutoh, H.; Knöpfel, T. Exploration of fluorescent protein voltage probes based on circularly permuted fluorescent proteins. Front. Neuroeng. 2009, 2. [Google Scholar] [CrossRef] [PubMed]
- Perron, A.; Mutoh, H.; Launey, T.; Knöpfel, T. Red-Shifted Voltage-Sensitive Fluorescent Proteins. Chem. Biol. 2009, 16, 1268–1277. [Google Scholar] [CrossRef] [PubMed]
- Akemann, W.; Mutoh, H.; Perron, A.; Park, Y.K.; Iwamoto, Y.; Knöpfel, T. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol. 2012, 108, 2323–2337. [Google Scholar] [CrossRef] [PubMed]
- Villalba-Galea, C.A.; Sandtner, W.; Dimitrov, D.; Mutoh, H.; Knöpfel, T.; Bezanilla, F. Charge movement of a voltage-sensitive fluorescent protein. Biophys. J. 2009, 96, L19–L21. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Jin, L.; Han, Z.; Cohen, L.B.; Popovic, M.; Platisa, J.; Pieribone, V. Genetically encoded fluorescent voltage sensors using the voltage-sensing domain of Nematostella and Danio phosphatases exhibit fast kinetics. J. Neurosci. Methods 2012, 208, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Jin, L.; Platisa, J.; Cohen, L.B.; Baker, B.J.; Pieribone, V.A. Fluorescent protein voltage probes derived from ArcLight that respond to membrane voltage changes with fast kinetics. PLoS ONE 2013, 8, e81295. [Google Scholar] [CrossRef] [PubMed]
- Kralj, J.M.; Hochbaum, D.R.; Douglass, A.D.; Cohen, A.E. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science 2011, 333, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Ernst, O.P.; Sánchez Murcia, P.A.; Daldrop, P.; Tsunoda, S.P.; Kateriya, S.; Hegemann, P. Photoactivation of channelrhodopsin. J. Biol. Chem. 2008, 283, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Li, J.Z.; Schnitzer, M.J. Enhanced Archaerhodopsin Fluorescent Protein Voltage Indicators. PLoS ONE 2013, 8, e66959. [Google Scholar] [CrossRef] [PubMed]
- Flytzanis, N.C.; Bedbrook, C.N.; Chiu, H.; Engqvist, M.K.M.; Xiao, C.; Chan, K.Y.; Sternberg, P.W.; Arnold, F.H.; Gradinaru, V. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.; Fromherz, P.; Denk, W. High Sensitivity of Stark-Shift Voltage-Sensing Dyes by One- or Two-Photon Excitation Near the Red Spectral Edge. Biophys. J. 2004, 87, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Bublitz, G.; King, B.; Boxer, S. Electronic structure of the Chromophore in Green Fluorescent Protein. J. Am. Chem. Soc. 1998, 120, 9370–9371. [Google Scholar] [CrossRef]
- Rosell, F.I.; Boxer, S.G. Polarized absorption spectra of green fluorescent protein single crystals: Transition dipole moment directions. Biochemistry 2003, 42, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Khatchatouriants, A.; Lewis, A.; Rothman, Z.; Loew, L.; Treinin, M. GFP is a selective non-linear optical sensor of electrophysiological processes in Caenorhabditis elegans. Biophys. J. 2000, 79, 2345–2352. [Google Scholar] [CrossRef]
- Kaestner, L.; Tian, Q.; Lipp, P. Action potentials in heart cells. In Fluorescent Proteins II; Jung, G., Ed.; Springer: Heidelberg, Germany, 2012; pp. 163–182. [Google Scholar]
- Kaestner, L.; Lipp, P. Screening action potentials: The power of light. Front. Pharmacol. 2011, 2, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Lipp, P.; Kaestner, L. Cardiac action potential imaging. In Progress in Biomedical Optics and Imaging—Proceedings of SPIE 8798; The Society of Photo-Optical Instrumentation Engineers (SPIE): Bellingham, WA, USA, 2013. [Google Scholar]
- Mutoh, H.; Knöpfel, T. Probing neuronal activities with genetically encoded optical indicators: From a historical to a forward-looking perspective. Pflügers Arch. 2013, 465, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Quintana, A.; Schwarz, E.C.; Schwindling, C.; Lipp, P.; Kaestner, L.; Hoth, M. Sustained activity of CRAC channels requires translocation of mitochondria to the plasma membrane. J. Biol. Chem. 2006, 281, 40302–40309. [Google Scholar] [CrossRef] [PubMed]
- Minetti, G.; Egée, S.; Mörsdorf, D.; Steffen, P.; Makhro, A.; Achilli, C.; Ciana, A.; Wang, J.; Bouyer, G.; Bernhardt, I.; et al. Red cell investigations: Art and artefacts. Blood Rev. 2013, 27, 91–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldwin, A.L.; Thurston, G. Mechanics of endothelial cell architecture and vascular permeability. Crit. Rev. Biomed. Eng. 2001, 29, 247–278. [Google Scholar] [CrossRef] [PubMed]
- Haugland, R.P. Handbook of Fluorescent Probes and Research Products, 9th ed.; Molecular Probes: Eugene, OR, USA, 2002. [Google Scholar]
- Kaestner, L.; Tian, Q.; Lipp, P. Cardiac safety screens: Molecular, cellular and optical advancements. In Progress in Biomedical Optics and Imaging—Proceedings of SPIE 8089; The Society of Photo-Optical Instrumentation Engineers (SPIE): Bellingham, WA, USA, 2011. [Google Scholar]
- Zeug, A.; Woehler, A.; Neher, E.; Ponimaskin, E.G. Quantitative Intensity-Based FRET Approaches—A Comparative Snapshot. Biophys. J. 2012, 103, 1821–1827. [Google Scholar] [CrossRef] [PubMed]
- Arrigoni, C.; Crivori, P. Assessment of QT liabilities in drug development. Cell Biol. Toxicol. 2007, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sinnecker, D.; Dirschinger, R.J.; Goedel, A.; Moretti, A.; Lipp, P.; Laugwitz, K.-L. Induced Pluripotent Stem Cells in Cardiovascular Research. Rev. Physiol. Biochem. Pharmacol. 2012, 163, 1–26. [Google Scholar] [PubMed]
- Matsa, E.; Burridge, P.W.; Wu, J.C. Human stem cells for modeling heart disease and for drug discovery. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Reil, J.C.; Hohl, M.; Oberhofer, M.; Kazakov, A.; Kaestner, L.; Mueller, P.; Adam, O.; Maack, C.; Lipp, P.; Mewis, C.; et al. Cardiac Rac1 overexpression in mice creates a substrate for atrial arrhythmias characterized by structural remodelling. Cardiovasc. Res. 2010, 87, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Yan, P.; Ewart, P.; Kohl, P.; Loew, L.M.; Bollensdorff, C. Simultaneous measurement and modulation of multiple physiological parameters in the isolated heart using optical techniques. Pflugers Arch. 2012, 464, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Acker, C.D.; Zhou, W.-L.; Lee, P.; Bollensdorff, C.; Negrean, A.; Lotti, J.; Sacconi, L.; Antic, S.D.; Kohl, P.; et al. Palette of fluorinated voltage-sensitive hemicyanine dyes. Proc. Natl. Acad. Sci. USA 2012, 109, 20443–20448. [Google Scholar] [CrossRef] [PubMed]
- Tallini, Y.N.; Ohkura, M.; Choi, B.R.; Ji, G.; Imoto, K.; Doran, R.; Lee, J.S.; Plan, P.; Wilson, J.; Xin, H.B.; et al. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. USA 2006, 103, 4753–4758. [Google Scholar] [CrossRef] [PubMed]
- Kaestner, L.; Scholz, A.; Tian, Q.; Ruppenthal, S.; Tabellion, W.; Wiesen, K.; Katus, H.A.; Müller, O.J.; Kotlikoff, M.I.; Lipp, P. Genetically encoded Ca2+ indicators in cardiac myocytes. Circ. Res. 2014, 114, 1623–1639. [Google Scholar] [CrossRef] [PubMed]
- Bers, D.M. Cardiac excitation-contraction coupling. Nature 2002, 415, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.; Fagerholm, E.D.; Mutoh, H.; Leech, R.; Sharp, D.J.; Shew, W.L.; Knöpfel, T. Voltage imaging of waking mouse cortex reveals emergence of critical neuronal dynamics. J. Neurosci. 2014, 34, 16611–16620. [Google Scholar] [CrossRef] [PubMed]
- Kaestner, L.; Scholz, A.; Lipp, P. Conceptual and technical aspects of transfection and gene delivery. Bioorg. Med. Chem. Lett. 2015, 25, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Hui, X.; Reither, G.; Kaestner, L.; Lipp, P. Targeted activation of conventional and novel protein kinase C through differential translocation patterns. Mol. Cell. Biol. 2014, 34, 2370–2381. [Google Scholar] [CrossRef] [PubMed]
- Kaestner, L.; Tabellion, W.; Weiss, E.; Bernhardt, I.; Lipp, P. Calcium imaging of individual erythrocytes: Problems and approaches. Cell Calcium 2006, 39, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Kaestner, L.; Lipp, P. Noise-free visualization of microscopic calcium signaling by pixel-wise fitting. Circ. Res. 2012, 111, 17–27. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaestner, L.; Tian, Q.; Kaiser, E.; Xian, W.; Müller, A.; Oberhofer, M.; Ruppenthal, S.; Sinnecker, D.; Tsutsui, H.; Miyawaki, A.; et al. Genetically Encoded Voltage Indicators in Circulation Research. Int. J. Mol. Sci. 2015, 16, 21626-21642. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160921626
Kaestner L, Tian Q, Kaiser E, Xian W, Müller A, Oberhofer M, Ruppenthal S, Sinnecker D, Tsutsui H, Miyawaki A, et al. Genetically Encoded Voltage Indicators in Circulation Research. International Journal of Molecular Sciences. 2015; 16(9):21626-21642. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160921626
Chicago/Turabian StyleKaestner, Lars, Qinghai Tian, Elisabeth Kaiser, Wenying Xian, Andreas Müller, Martin Oberhofer, Sandra Ruppenthal, Daniel Sinnecker, Hidekazu Tsutsui, Atsushi Miyawaki, and et al. 2015. "Genetically Encoded Voltage Indicators in Circulation Research" International Journal of Molecular Sciences 16, no. 9: 21626-21642. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160921626