Identification of Human UDP-Glucuronosyltransferase 1A4 as the Major Isozyme Responsible for the Glucuronidation of 20(S)-Protopanaxadiol in Human Liver Microsomes
Abstract
:1. Introduction
2. Results
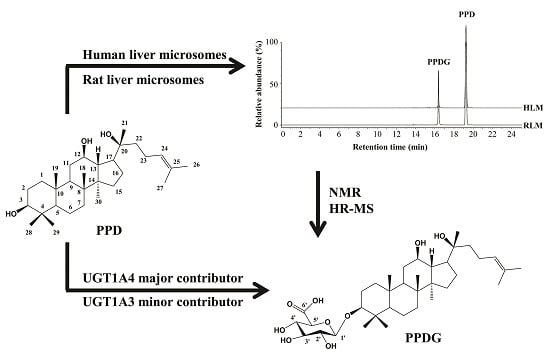
2.1. Identification and Structural Elucidation of Metabolite



| Position | 13C NMR | 1H NMR | ||
|---|---|---|---|---|
| PPD a | PPDG | PPD a | PPDG | |
| 1 | 39.4, t | 39.0,t | - | - |
| 2 | 28.2, t | 29.4, t | - | - |
| 3 | 78.4, d | 90.1, d | - | - |
| 4 | 39.5, s | 38.6, s | - | - |
| 5 | 56.4, d | 56.1, d | - | - |
| 6 | 18.8, d | 17.8, d | - | - |
| 7 | 35.2, t | 34.6, t | - | - |
| 8 | 40.0, d | 39.6, d | - | - |
| 9 | 50.5, d | 50.0, d | - | - |
| 10 | 37.4, s | 36.6, s | - | - |
| 11 | 32.1, t | 31.7, t | - | - |
| 12 | 71.0, d | 71.9, d | 3.9, m | 4.1, m |
| 13 | 48.6, d | 48.2, d | - | - |
| 14 | 51.7, s | 51.2, s | - | - |
| 15 | 31.4, t | 30.7, t | - | - |
| 16 | 26.8, t | 26.0, t | - | - |
| 17 | 54.8, d | 53.7, d | 2.33, J = 10.7, 7.1 b | 2.37, dd, J = 15, 7.8 |
| 18 | 15.9, q | 15.4, q | 0.99, s | 1.06, s |
| 19 | 16.4, q | 15.7, q | 0.87, s, | 0.87, s |
| 20 | 72.9, s | 74.0, s | - | - |
| 21 | 27.1, q | 27.0, q | 1.41, s | 1.31, s |
| 22 | 35.9, t | 35.0, t | - | - |
| 23 | 23.0, t | 24.5, t | 2.28, s | - |
| 24 | 126.3, d | 125.9, d | 5.3, J = 7.1 | 5.1, dd, J = 7.6, 1.2 |
| 25 | 130.7, s | 130.6, s | - | - |
| 26 | 25.8, q | 25.9, q | 1.64, s | 1.70, s |
| 27 | 17.7, q | 15.7, q | 1.61, s | 1.64, s |
| 28 | 28.7, q | 29.1, q | 1.21, s | 1.16, s |
| 29 | 16.3, q | 14.8, q | 1.02, s | 1.03, s |
| 30 | 17.0, q | 15.4, q | 0.92, s | 0.94, s |
| Glucuronic acid | ||||
| 1′ | - | 106.4, d | - | 4.35, d, J = 7.8 |
| 2′ | - | 76.3, d | - | - |
| 3′ | - | 77.7, d | - | - |
| 4′ | - | 73.5, d | - | - |
| 5′ | - | 75.2, d | - | - |
| 6′ | - | 175.3, s | - | - |
2.2. Kinetics of 20(S)-Protopanaxadiol (PPD) Glucuronidation by Pooled Human Liver Microsomes (HLMs) and Rat Liver Microsomes (RLMs)

| Species | Vmax (nmol/min/mg Protein) | S50 (μM) | n | CLmax (μL/min/mg Protein) | R2 |
|---|---|---|---|---|---|
| RLMs | 0.30 ± 0.01 | 20.68 ± 1.39 | 2.50 ± 0.48 | 7.36 ± 0.27 | 0.9793 |
| HLMs | 0.32 ± 0.01 | 42.80 ± 0.73 | 2.12 ± 0.24 | 3.70 ± 0.01 | 0.9753 |
| UGT1A4 | 0.31 ± 0.01 | 18.49 ± 1.85 | 2.34 ± 0.23 | 8.39 ± 0.79 | 0.9894 |
2.3. Chemical Inhibition in Pooled HLMs

2.4. Assay with Recombinant Human UGTs

2.5. Kinetics of PPD Glucuronidation by Recombinant UGT1A4
3. Discussion
4. Experimental Section
4.1. Materials
4.2. Glucuronidation of PPD with the Pooled HLMs and RLMs
4.3. Identification of the Metabolite by UPLC-Q/TOF-MS
4.4. Preparation and Structural Elucidation of Metabolite
4.5. Quantitative Analysis by LC-MS/MS
4.6. Chemical Inhibition in Pooled HLMs
4.7. Incubation with Recombinant Human UGTs
4.8. Kinetics of PPD Glucuronidation in Recombinant UGT 1A4
4.9. Enzyme Kinetic Data Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hao, H.; Lai, L.; Zheng, C.; Wang, Q.; Yu, G.; Zhou, X.; Wu, L.; Gong, P.; Wang, G. Microsomal cytochrome p450-mediated metabolism of protopanaxatriol ginsenosides: Metabolite profile, reaction phenotyping, and structure-metabolism relationship. Drug Metab. Dispos. 2010, 38, 1731–1739. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, X.; Li, D.; Zhong, D. Identification of 20(S)-protopanaxadiol metabolites in human liver microsomes and human hepatocytes. Drug Metab. Dispos. 2011, 39, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, G.; Haitang, X.; Hao, L.; Guoyu, P.; Tucker, I. Simultaneous rapid quantification of ginsenoside Rg1 and its secondary glycoside Rh1 and aglycone protopanaxatriol in rat plasma by liquid chromatography–mass spectrometry after solid-phase extraction. J. Pharma. Biomed. Anal. 2005, 38, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Z.; Kim, K.E.; Du, G.J.; Qi, L.W.; Wen, X.D.; Li, P.; Bauer, B.A.; Bissonnette, M.B.; Musch, M.W.; Chang, E.B. Ultra-performance liquid chromatography and time-of-flight mass spectrometry analysis of ginsenoside metabolites in human plasma. Am. J. Chin. Med. 2011, 39, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M.; Kim, S.D.; Kim, K.S.; Song, Y.B.; Kwak, Y.S.; Cho, J.Y.; Park, H.J.; Oh, J.W.; Rhee, M.H. Protopanaxadiol modulates LPS-induced inflammatory activity in murinemacrophage RAW264.7 cells. J. Ginseng Res. 2006, 30, 181–187. [Google Scholar]
- Hui, Y.; Yang, Z.; Yang, Z.; Ge, Q. Inventors; CN-knowhow intellectual property agent limited, assignee. Antidepressant Compos. Contain. 2007, 20. [Google Scholar]
- Li, G.; Wang, Z.; Sun, Y.; Liu, K.; Wang, Z. Ginsenoside 20(S)-protopanaxadiol inhibits the proliferation and invasion of human fibrosarcoma HT1080 cells. Basic Clin. Pharmacol. 2006, 98, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Bu, X.; Yan, H.; Jia, W.W.G. 20S-protopanaxadiol-induced programmed cell death in glioma cells through caspase-dependent and-independent pathways. J. Nat. Prod. 2007, 70, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, H.; Yu, X.; Qu, S.; Chen, M.; Sui, D. Effect of 20(S)-protopanaxadiol on SMMC-7721 human liver cancer in vivo and in vitro. Chin. Pharmacol. Bull. 2008, 24, 1504–1508. [Google Scholar]
- Qi, L.W.; Wang, C.Z.; Yuan, C.S. American ginseng: Potential structure–function relationship in cancer chemoprevention. Biochem. Pharmacol. 2010, 80, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.Y.; Li, Y.W.; Tse, A.K.W.; Hau, D.K.P.; Leung, C.H.; Yu, Z.L.; Fong, W.F. 20(S)-protopanaxadiol, a metabolite of ginsenosides, induced cell apoptosis through endoplasmic reticulum stress in human hepatocarcinoma HepG2 cells. Eur. J. Pharmacol. 2011, 668, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Teng, J.; Chen, W.; Ge, Q.; Yang, Z.; Yu, C.; Yang, Z.; Jia, W. 20(S)-protopanaxadiol, an active ginseng metabolite, exhibits strong antidepressant-like effects in animal tests. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2010, 34, 1402–1411. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Wang, Q.; Tang, P. Lithium adduct as precursor ion for sensitive and rapid quantification of 20(S)-protopanaxadiol in rat plasma by liquid chromatography/quadrupole linear ion trap mass spectrometry and application to rat pharmacokinetic study. J. Mass Spectrom. 2013, 48, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Li, S.L.; Zhang, Z.H.; Zhu, F.X.; Sun, E.; Wei, Y.J.; Jia, X.B. Characterization of metabolites of 20(S)-protopanaxadiol in rats using ultra-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. J. Chromatogr. B 2013, 933, 59–66. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Li, J.; Wang, R.; Li, Z.; Annie Bligh, S.; Yang, L.; Wang, Z. Metabolic profiles of 20(S)-protopanaxadiol in rats after oral administration using ultra-performance liquid chromatography/quadrupole time-of-flight tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2014, 28, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Kiang, T.K.; Ensom, M.H.; Chang, T.K.H. UDP-glucuronosyltransferases and clinical drug–drug interactions. Pharmacol. Ther. 2005, 106, 97–132. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.G.; Jiang, Y.T.; Song, S.J.; Wang, Z.H.; Bai, J.; Xu, S.X.; Liu, K. Alkaline-degradation products of ginsenosides from leaves and stems of Panax quinquefolium. Acta Pharm. Sin. 2005, 40, 924–930. [Google Scholar]
- Cai, B.X.; Li, X.Y.; Chen, J.H.; Tang, Y.B.; Wang, G.L.; Zhou, J.G.; Qui, Q.Y.; Guan, Y.Y. Ginsenoside-Rd, a new voltage-independent Ca2+ entry blocker, reverses basilar hypertrophic remodeling in stroke-prone renovascular hypertensive rats. Eur. J. Pharmacol. 2009, 606, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.F.; Yang, J.L.; Du, F.F.; Gao, X.M.; Ma, X.T.; Huang, Y.H.; Xu, F.; Niu, W.; Wang, F.Q.; Mao, Y.; et al. Absorption and disposition of ginsenosides afteroral administration of Panax notoginseng extract to rats. Drug Metab. Dispos. 2009, 37, 2290–2298. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, J.; Liu, X.; Fu, Y.; Zhang, M.; Lin, Q.; Zhu, J.; Mai, L.; Shan, Z.; Yu, X.; et al. Panax notoginseng saponins inhibit ischemia-induced apoptosis by activating PI3K/Akt pathway in cardiomyocytes. J. Ethnopharmacol. 2011, 137, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Wang, M.; Venema, K.; Maathuis, A.; Heijden, R.; Greef, J.; Xu, G.W.; Hankemeier, T. Bioconversion of red ginseng saponins in the gastro-intestinal tract in vitro model studied by high-performance liquid chromatography–high resolution Fourier transform ion cyclotron resonance mass spectrometry. J. Chromatogr. A 2009, 1216, 2195–2203. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Hua, H.Y.; Liu, X.Y.; Liu, J.H.; Yu, B.Y. In vitro biotransformation of red ginseng extract by human intestinal microflora: Metabolites identification and metabolic profile elucidation using LC–Q-TOF/MS. J. Pharm. Biomed. 2014, 98, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, T.D.; Callaghan, J.T.; Einolf, H.J.; Fischer, V.; Gan, L.; Grimm, S.; Kao, J.; King, S.P.; Miwa, G. The conduct of in vitro and in vivo drug–drug interaction studies: A Pharmaceutical Research and Manufacturers of America (PhRMA) perspective. Drug Metab. Dispos. 2003, 31, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; He, Y.Q.; Hu, Y.; Liu, Y.; Zhang, J.W.; Li, W.; Wang, Z.T.; Yang, L. Determination of UDP-glucuronosyltransferase UGT2B7 activity in human liver microsomes by ultra-performance liquid chromatography with MS detection. J. Chromatogr. B 2008, 870, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Uchaipichat, V.; Mackenzie, P.I.; Elliot, D.J.; Miners, J.O. Selectivity of substrate (trifluoperazine) and inhibitor (amitriptyline, androsterone, canrenoic acid, hecogenin, phenylbutazone, quinidine, quinine, and sulfinpyrazone) “probes” for human UDP-glucuronosyltransferases. Drug Metab. Dispos. 2006, 34, 449–456. [Google Scholar] [PubMed]
- Soars, M.G.; Ring, B.J.; Wrighton, S.A. The effect of incubation conditions on the enzyme kinetics of UDP-glucuronosyltransferases. Drug Metab. Dispos. 2003, 31, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Senafi, S.B.; Clarke, D.J.; Burchell, B. Investigation of the substrate specificity of a cloned expressed human bilirubin UDP-glucuronosyltransferase: UDP-sugar specificity and involvementin steroid and xenobiotic glucuronidation. Biochem. J. 1994, 303, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Gall, W.E.; Zawada, G.; Mojarrabi, B.; Tephly, T.R.; Green, M.D.; Coffman, B.L.; Mackenzie, P.I.; Radominska, P.A. Differential glucuronidation of bile acids androgens and estrogens by human UGT1A3 and 2B7. J. Steroid Biochem. Mol. Biol. 1999, 70, 101–108. [Google Scholar] [CrossRef]
- Soars, M.G.; Petullo, D.M.; Eckstein, J.A.; Kasper, S.C.; Wrighton, S.A. An assessment of UDP-glucuronosyltransferase induction using primary human hepatocytes. Drug Metab. Dispos. 2004, 32, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.W.; Liu, Y.; Zhao, J.Y.; Wang, L.M.; Ge, G.B.; Gao, Y.; Li, W.; Liu, H.T.; Liu, H.X.; Zhang, Y.Y. Metabolic profiling and cytochrome p450 reaction phenotyping of medroxyprogesterone acetate. Drug Metab. Dispos. 2008, 36, 2292–2298. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhou, D.; Li, J.; Han, H.; Ji, G.; Yang, L.; Wang, Z. Identification of 20(S)-protopanaxatriol metabolites in rats by ultra-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry and nuclear magnetic resonance spectroscopy. J. Pharm. Biomed. 2014, 88, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Liu, Y.; Zhang, J.W.; Li, W.; Liu, H.T.; Yang, L. UDP-glucuronosyltransferase 1A6 is the major isozyme responsible for protocatechuic aldehyde glucuronidation in human liver microsomes. Drug Metab. Dispos. 2008, 36, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Tallman, M.N.; Ali, S.Y.; Smith, P.C. UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human liver and intestinal microsomes: Structural characterization of phenolic and alcoholic glucuronides of etoposide and estimation of enzyme kinetics. Drug Metab. Dispos. 2007, 35, 371–380. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Q.; Liu, Y.; Zhang, B.F.; Liu, H.X.; Lu, Y.L.; Yang, L.; Xiong, A.Z.; Xu, L.L.; Wang, C.H.; Yang, L. Identification of the UDP-glucuronosyltransferase isozyme involved in senecionine glucuronidation in human liver microsomes. Drug Metab. Dispos. 2010, 38, 626–634. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; He, C.; Fang, L.; Yang, L.; Wang, Z. Identification of Human UDP-Glucuronosyltransferase 1A4 as the Major Isozyme Responsible for the Glucuronidation of 20(S)-Protopanaxadiol in Human Liver Microsomes. Int. J. Mol. Sci. 2016, 17, 205. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030205
Li J, He C, Fang L, Yang L, Wang Z. Identification of Human UDP-Glucuronosyltransferase 1A4 as the Major Isozyme Responsible for the Glucuronidation of 20(S)-Protopanaxadiol in Human Liver Microsomes. International Journal of Molecular Sciences. 2016; 17(3):205. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030205
Chicago/Turabian StyleLi, Jia, Chunyong He, Lianxiang Fang, Li Yang, and Zhengtao Wang. 2016. "Identification of Human UDP-Glucuronosyltransferase 1A4 as the Major Isozyme Responsible for the Glucuronidation of 20(S)-Protopanaxadiol in Human Liver Microsomes" International Journal of Molecular Sciences 17, no. 3: 205. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030205