Transcriptomic Insights into the Response of Placenta and Decidua Basalis to the CpG Oligodeoxynucleotide Stimulation in Non-Obese Diabetic Mice and Wild-Type Controls
Abstract
:1. Introduction
2. Results
2.1. Effects of CpG ODN on Embryo Loss
2.2. Illumina Sequencing and Gene Expression Profiles
2.3. Gene Ontology and Pathway Analysis
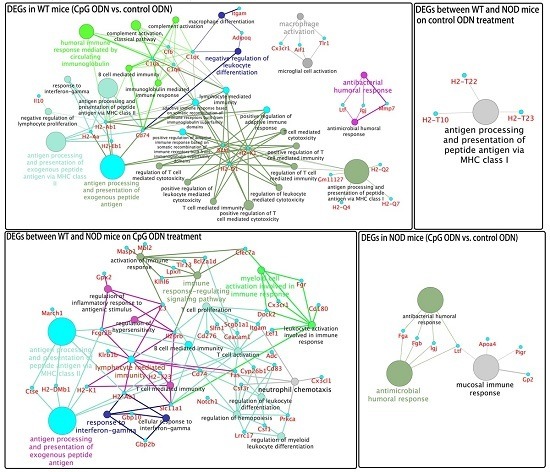
2.4. Functional Groups in Immune System Processes
2.5. Validation of Gene Expression
2.6. Macrophage Polarization
2.7. Effect of C1q Inhibition on Abortion in Wild-Type (WT) Mice
3. Discussion
4. Materials and Methods
4.1. Animal Administration and Sample Collection
4.2. RNA Preparation and Construction of RNA-Seq Libraries
4.3. Sequencing and Assembly
4.4. Data Analysis
4.5. Quantitative Real-Time PCR
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Erlebacher, A. Immunology of the maternal-fetal interface. Annu. Rev. Immunol. 2013, 31, 387–411. [Google Scholar]
- Salker, M.; Teklenburg, G.; Molokhia, M.; Lavery, S.; Trew, G.; Aojanepong, T.; Mardon, H.J.; Lokugamage, A.U.; Rai, R.; Landles, C.; et al. Natural selection of human embryos: Impaired decidualization of endometrium disables embryo-maternal interactions and causes recurrent pregnancy loss. PLoS ONE 2010, 5, e10287. [Google Scholar]
- Romero, R.; Espinoza, J.; Mazor, M. Can endometrial infection/inflammation explain implantation failure, spontaneous abortion, and preterm birth after in vitro fertilization? Fertil. Steril. 2004, 82, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002, 20, 709–760. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.M.; Yi, A.K.; Matson, S.; Waldschmidt, T.J.; Bishop, G.A.; Teasdale, R.; Koretzky, G.A.; Klinman, D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 1995, 374, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, G.; Andresen, L.; Matthiessen, M.W.; Rask-Madsen, J.; Brynskov, J. Expression of toll-like receptor 9 and response to bacterial CpG oligodeoxynucleotides in human intestinal epithelium. Clin. Exp. Immunol. 2005, 141, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Thaxton, J.E.; Romero, R.; Sharma, S. TLR9 activation coupled to IL-10 deficiency induces adverse pregnancy outcomes. J. Immunol. 2009, 183, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Qin, X.; Shan, B.; Wang, W.; Zhu, Q.; Sharma, S.; Wu, J.; Lin, Y. Differential effects of the CpG-Toll-like receptor 9 axis on pregnancy outcome in nonobese diabetic mice and wild-type controls. Fertil. Steril. 2013, 99, 1759–1767. [Google Scholar] [CrossRef] [PubMed]
- Formby, B.; Schmid-Formby, F.; Jovanovic, L.; Peterson, C.M. The offspring of the female diabetic “nonobese diabetic” (NOD) mouse are large for gestational age and have elevated pancreatic insulin content: A new animal model of human diabetic pregnancy. In Proceedings of the Society for Experimental Biology and Medicine; Society for Experimental Biology and Medicine: New York, NY, USA, 1987; Volume 184, pp. 291–294. [Google Scholar]
- Kim, J.H.; Bugaj, L.J.; Oh, Y.J.; Bivalacqua, T.J.; Ryoo, S.; Soucy, K.G.; Santhanam, L.; Webb, A.; Camara, A.; Sikka, G.; et al. Arginase inhibition restores NOS coupling and reverses endothelial dysfunction and vascular stiffness in old rats. J. Appl. Physiol. 2009, 107, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Mehl, A.; Ghorbani, P.; Douda, D.; Huang, H.; Palaniyar, N.; Ratjen, F.; Grasemann, H. Effect of arginase inhibition on pulmonary L-arginine metabolism in murine Pseudomonas pneumonia. PLoS ONE 2014, 9, e90232. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, X.; Shan, B.; Wu, J.; Sharma, S.; Sun, Y. Prevention of CpG-induced pregnancy disruption by adoptive transfer of in vitro-induced regulatory T cells. PLoS ONE 2014, 9, e94702. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ren, L.; Wang, W.; Di, J.; Zeng, S.; Saito, S. Effect of TLR3 and TLR7 activation in uterine NK cells from non-obese diabetic (NOD) mice. J. Reprod. Immunol. 2009, 82, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D. M1 and M2 Macrophages: Oracles of Health and Disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [PubMed]
- Bohlson, S.S.; O’Conner, S.D.; Hulsebus, H.J.; Ho, M.M.; Fraser, D.A. Complement, C1q, and C1q-related molecules regulate macrophage polarization. Front. Immunol. 2014, 5, 402. [Google Scholar] [CrossRef] [PubMed]
- Benoit, M.E.; Clarke, E.V.; Morgado, P.; Fraser, D.A.; Tenner, A.J. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J. Immunol. 2012, 188, 5682–5693. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Kirschning, C.J.; Hacker, H.; Redecke, V.; Hausmann, S.; Akira, S.; Wagner, H.; Lipford, G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 2001, 98, 9237–9242. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.T. Lactoferrin gene expression and regulation: An overview. Biochem. Cell Biol. 2002, 80, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Otsuki, K.; Tokunaka, M.; Oba, T.; Nakamura, M.; Shirato, N.; Okai, T. Administration of oral and vaginal prebiotic lactoferrin for a woman with a refractory vaginitis recurring preterm delivery: Appearance of lactobacillus in vaginal flora followed by term delivery. J. Obstet. Gynaecol. Res. 2014, 40, 583–585. [Google Scholar] [CrossRef] [PubMed]
- Paesano, R.; Pietropaoli, M.; Berlutti, F.; Valenti, P. Bovine lactoferrin in preventing preterm delivery associated with sterile inflammation. Biochem. Cell Biol. 2012, 90, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, Y.; Otsuki, K.; Yoda, A.; Shimizu, Y.; Saito, H.; Yanaihara, T. Effect of lactoferrin on lipopolysaccharide (LPS) induced preterm delivery in mice. Acta Obstet. Gynecol. Scand. 2000, 79, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Giunta, G.; Giuffrida, L.; Mangano, K.; Fagone, P.; Cianci, A. Influence of lactoferrin in preventing preterm delivery: A pilot study. Mol. Med. Rep. 2012, 5, 162–166. [Google Scholar] [PubMed]
- Hasegawa, A.; Otsuki, K.; Sasaki, Y.; Sawada, M.; Mitsukawa, K.; Chiba, H.; Nagatsuka, M.A.; Okai, T.; Kato, A. Preventive effect of recombinant human lactoferrin in a rabbit preterm delivery model. Am. J. Obstet. Gynecol. 2005, 192, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Otsuki, K.; Hasegawa, A.; Sawada, M.; Chiba, H.; Negishi, M.; Nagatsuka, M.; Okai, T. Preventive effect of recombinant human lactoferrin on lipopolysaccharide-induced preterm delivery in mice. Acta Obstet. Gynecol. Scand. 2004, 83, 1035–1038. [Google Scholar] [CrossRef] [PubMed]
- Otsuki, K.; Yakuwa, K.; Sawada, M.; Hasegawa, A.; Sasaki, Y.; Mitsukawa, K.; Chiba, H.; Nagatsuka, M.; Saito, H.; Okai, T. Recombinant human lactoferrin has preventive effects on lipopolysaccharide-induced preterm delivery and production of inflammatory cytokines in mice. J. Perinat. Med. 2005, 33, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Otsuki, K.; Mitsuhashi, Y.; Yoda, A.; Yanaihara, A.; Shimizu, Y.; Yanaihara, T. Role of lactoferrin in the prevention of preterm delivery. Int. Congr. Ser. 2000, 1195, 347–352. [Google Scholar]
- Teng, C.T.; Beard, C.; Gladwell, W. Differential expression and estrogen response of lactoferrin gene in the female reproductive tract of mouse, rat, and hamster. Biol. Reprod. 2002, 67, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Yakuwa, K.; Otsuki, K.; Nakayama, K.; Hasegawa, A.; Sawada, M.; Mitsukawa, K.; Chiba, H.; Nagatsuka, M.; Okai, T. Recombinant human lactoferrin has a potential to suppresses uterine cervical ripening in preterm delivery in animal model. Arch. Gynecol. Obstet. 2007, 275, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Locci, M.; Nazzaro, G.; Miranda, M.; Salzano, E.; Montagnani, S.; Castaldo, C.; De Placido, G. Vaginal lactoferrin in asymptomatic patients at low risk for pre-term labour for shortened cervix: Cervical length and interleukin-6 changes. J. Obstet. Gynaecol. 2013, 33, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Koshland, M.E. The coming of age of the immunoglobulin J chain. Annu. Rev. Immunol. 1985, 3, 425–453. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, B.A.; Conner, D.A.; Ladd, D.J.; Kendall, D.; Casanova, J.E.; Corthesy, B.; Max, E.E.; Neutra, M.R.; Seidman, C.E.; Seidman, J.G. Altered hepatic transport of immunoglobulin A in mice lacking the J chain. J. Exp. Med. 1995, 182, 1905–1911. [Google Scholar] [CrossRef] [PubMed]
- Johansen, F.E.; Braathen, R.; Brandtzaeg, P. Role of J chain in secretory immunoglobulin formation. Scand. J. Immunol. 2000, 52, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Vaerman, J.P.; Langendries, A.E.; Giffroy, D.A.; Kaetzel, C.S.; Fiani, C.M.; Moro, I.; Brandtzaeg, P.; Kobayashi, K. Antibody against the human J chain inhibits polymeric Ig receptor-mediated biliary and epithelial transport of human polymeric IgA. Eur. J. Immunol. 1998, 28, 171–182. [Google Scholar] [CrossRef]
- Tacchi, L.; Larragoite, E.; Salinas, I. Discovery of J chain in African lungfish (Protopterus dolloi, Sarcopterygii) using high throughput transcriptome sequencing: Implications in mucosal immunity. PLoS ONE 2013, 8, e70650. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.M.; Wolf, D.; Rumpold, H.; Enrich, B.; Tilg, H. Adiponectin induces the anti-inflammatory cytokines IL-10 and IL-1RA in human leukocytes. Biochem. Biophys. Res. Commun. 2004, 323, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Argueta, J.G.; Masuhiro, Y.; Kagishita, M.; Nonaka, K.; Saito, T.; Hanazawa, S.; Yamashita, Y. Adiponectin inhibits Toll-like receptor family-induced signaling. FEBS Lett. 2005, 579, 6821–6826. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Oritani, K.; Takahashi, I.; Ishikawa, J.; Matsuyama, A.; Ouchi, N.; Kihara, S.; Funahashi, T.; Tenner, A.J.; Tomiyama, Y.; et al. Adiponectin, a new member of the family of soluble defense collagens, negatively regulates the growth of myelomonocytic progenitors and the functions of macrophages. Blood 2000, 96, 1723–1732. [Google Scholar] [PubMed]
- Barnes, M.A.; Carson, M.J.; Nair, M.G. Non-traditional cytokines: How catecholamines and adipokines influence macrophages in immunity, metabolism and the central nervous system. Cytokine 2015, 72, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Bou, M.; Todorcevic, M.; Rodriguez, J.; Capilla, E.; Gutierrez, J.; Navarro, I. Interplay of adiponectin, TNFalpha and insulin on gene expression, glucose uptake and PPARγ, AKT and TOR pathways in rainbow trout cultured adipocytes. Gen. Comp. Endocrinol. 2014, 205, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Chinetti, G.; Zawadski, C.; Fruchart, J.C.; Staels, B. Expression of adiponectin receptors in human macrophages and regulation by agonists of the nuclear receptors PPARα, PPARγ, and LXR. Biochem. Biophys. Res. Commun. 2004, 314, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Geeraert, B.; Arnould, T.; Tsatsanis, C.; Holvoet, P. PPAR agonist-induced reduction of Mcp1 in atherosclerotic plaques of obese, insulin-resistant mice depends on adiponectin-induced Irak3 expression. PLoS ONE 2013, 8, e62253. [Google Scholar] [CrossRef] [PubMed]
- FASTX-Toolkit. Available online: URL http://hannonlab.cshl.edu/fastx_toolkit/ (accessed on 2 February 2010).
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trouw, L.A.; Seelen, M.A.; Duijs, J.M.; Benediktsson, H.; Van Kooten, C.; Daha, M.R. Glomerular deposition of C1q and anti-C1q antibodies in mice following injection of antimouse C1q antibodies. Clin. Exp. Immunol. 2003, 132, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007, 2, 2366–2382. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.-R.; Guo, Y.-N.; Qin, C.-M.; Qin, X.-L.; Tao, F.; Su, F.; Tian, F.-J.; Zhang, Y.; Lin, Y. Transcriptomic Insights into the Response of Placenta and Decidua Basalis to the CpG Oligodeoxynucleotide Stimulation in Non-Obese Diabetic Mice and Wild-Type Controls. Int. J. Mol. Sci. 2016, 17, 1281. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17081281
Liu X-R, Guo Y-N, Qin C-M, Qin X-L, Tao F, Su F, Tian F-J, Zhang Y, Lin Y. Transcriptomic Insights into the Response of Placenta and Decidua Basalis to the CpG Oligodeoxynucleotide Stimulation in Non-Obese Diabetic Mice and Wild-Type Controls. International Journal of Molecular Sciences. 2016; 17(8):1281. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17081281
Chicago/Turabian StyleLiu, Xiao-Rui, Yu-Na Guo, Chuan-Mei Qin, Xiao-Li Qin, Fei Tao, Fei Su, Fu-Ju Tian, Yan Zhang, and Yi Lin. 2016. "Transcriptomic Insights into the Response of Placenta and Decidua Basalis to the CpG Oligodeoxynucleotide Stimulation in Non-Obese Diabetic Mice and Wild-Type Controls" International Journal of Molecular Sciences 17, no. 8: 1281. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17081281