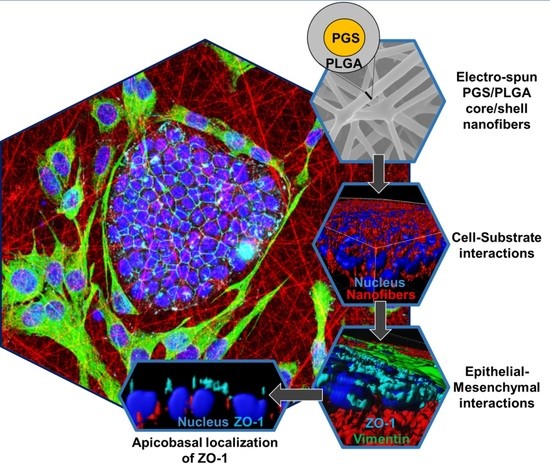
Mesenchymal Cells Affect Salivary Epithelial Cell Morphology on PGS/PLGA Core/Shell Nanofibers
Abstract
:1. Introduction
2. Results
2.1. Development and Characterization of PGS/PLGA Core/Shell Nanofibers
2.1.1. Electrospinning Parameter Optimization and SEM Fiber Morphology
2.1.2. Physical Properties of PGS/PLGA and PLGA Fiber Mats
2.2. SIMS Cell Growth on PGS/PLGA Nanofiber Substrates
2.3. Effect of Cell Morphology on Softer PGS/PLGA Fiber Mats
2.3.1. SIMS Cell Morphology on PGS/PLGA vs. PLGA Nanofiber Substrates
2.3.2. Epithelial–Mesenchymal Cell Self-Organization and Penetration into Scaffolds
2.3.3. SIMS Cell Arrangement and Morphology with Mesenchymal Cell Interactions
2.4. Cell–Cell Tight Junction Localization of SIMS in Mono- and Co-Culture Systems
3. Materials and Methods
3.1. Materials
3.2. Synthesis of Poly (Glycerol Sebacate) (PGS)
3.3. Nanofiber Fabrication
3.3.1. Fabrication of Nanofibrous PLGA Scaffolds
3.3.2. Fabrication of Nanofibrous PGS/PLGA Scaffolds
3.4. Nanofiber Characterization
3.4.1. Characterization of Fiber Morphology Using Scanning Electron Microscopy (SEM)
3.4.2. Material Characterization
3.5. Cell Culture
3.6. MTT Assay
3.7. Immunocytochemistry and Confocal Microscopy
3.8. Statistical Analysis
3.9. Bio-LIME: Quantitative Analysis for Cell Penetration Depth, Cell Morphology and Cell–Cell Junction Protein Localization Measurements
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yoo, C.; Vines, J.B.; Alexander, G.; Murdock, K.; Hwang, P.; Jun, H.-W. Adult stem cells and tissue engineering strategies for salivary gland regeneration: A review. Biomater. Res. 2014, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Aframian, D.J.; Cukierman, E.; Nikolovski, J.; Mooney, D.J.; Yamada, K.M.; Baum, B.J. The growth and morphological behavior of salivary epithelial cells on matrix protein-coated biodegradable substrata. Tissue Eng. 2000, 6, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.H.; Huang, T.W.; Chou, Y.S.; Hsu, S.H.; Su, W.F.; Lou, P.J.; Young, T.H. Formation of post-confluence structure in human parotid gland acinar cells on PLGA through regulation of E-cadherin. Biomaterials 2012, 3, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.A.; Larsen, M. Heterotypic control of basement membrane dynamics during branching morphogenesis. Dev. Biol. 2015, 401, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Gentile, P.; Chiono, V.; Carmagnola, I.; Hatton, P.V. An overview of poly(lactic-co-glycolic) acid (PLGA)-based biomaterials for bone tissue engineering. Int. J. Mol. Sci. 2014, 15, 3640–3659. [Google Scholar] [CrossRef] [PubMed]
- Milleret, V.; Simona, B.; Neuenschwander, P.; Hall, H. Tuning electrospinning parameters for production of 3D-fiber-fleeces with increased porosity for soft tissue engineering applications. Eur. Cell. Mater. 2011, 21, 286–303. [Google Scholar] [CrossRef] [PubMed]
- Stachewicz, U.; Qiao, T.; Rawlinson, S.C.F.; Almeida, F.V.; Cattell, W.-Q.; Li, M.A.H. Barber, 3D imaging of cell interactions with electrospun PLGA nanofiber membranes for bone regeneration. Acta Biomater. 2015, 27, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Mahjour, S.B.; Fu, X.; Yang, X.; Fong, J.; Sefat, F.; Wang, H. Rapid creation of skin substitutes from human skin cells and biomimetic nanofibers for acute full-thickness wound repair. Burns 2015, 41, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Said, S.S.; El-Halfawy, O.M.; El-Gowelli, H.M.; Aloufy, A.K.; Boraei, N.A.; El-Khordagui, L.K. Bioburden-responsive antimicrobial PLGA ultrafine fibers for wound healing. Eur. J. Pharm. Biopharm. 2012, 80, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.S.; Kim, T.G.; Park, T.G. Surface-functionalized electrospun nanofibers for tissue engineering and drug delivery. Adv. Drug Deliv. Rev. 2009, 61, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Cantara, S.I.; Soscia, D.A.; Sequeira, S.J.; Jean-Gilles, R.P.; Castracane, J.; Larsen, M. Selective functionalization of nanofiber scaffolds to regulate salivary gland epithelial cell proliferation and polarity. Biomaterials 2012, 33, 8372–8382. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M.; Van Itallie, C.M. Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 2009, 1, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Soscia, D.A.; Sequeira, S.J.; Schramm, R.A.; Jayarathanam, K.; Cantara, S.I.; Larsen, M.; Castracane, J. Salivary gland cell differentiation and organization on micropatterned PLGA nanofiber craters. Biomaterials 2013, 34, 6773–6784. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fan, X.; Zhu, J.; Xu, G.; Li, Y.; Liu, X. Neuroscience Letters Co-culturing improves the OGD-injured neuron repairing and NSCs differentiation via Notch pathway activation. Neurosci. Lett. 2014, 559, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, M.; Nikkhah, M.; Shin, S.-R.; Annabi, N.; Masoumi, N.; Gaharwar, A.K.; Camci-Unal, G.; Khademhosseini, A. PGS: Gelatin nanofibrous scaffolds with tunable mechanical and structural properties for engineering cardiac tissues. Biomaterials 2013, 34, 6355–6366. [Google Scholar] [CrossRef] [PubMed]
- Sill, T.J.; von Recum, H.A. Electrospinning: Applications in drug delivery and tissue engineering. Biomaterials 2008, 29, 1989–2006. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.B.; Nelson, D.A.; Kwon, H.R.; Koslow, M.; DeSantis, K.A.; Larsen, M. TGFβ signaling promotes matrix assembly during mechanosensitive embryonic salivary gland restoration. Matrix Biol. 2015, 43, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Mosier, A.P.; Peters, S.B.; Larsen, M.; Cady, N.C. Microfluidic Platform for the Elastic Characterization of Mouse Submandibular Glands by Atomic Force Microscopy. Biosensors 2014, 4, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liang, S.; Thouas, G.A. Elastomeric biomaterials for tissue engineering. Prog. Polym. Sci. 2013, 38, 584–671. [Google Scholar] [CrossRef]
- Patel, A.; Gaharwar, A.K.; Iviglia, G.; Zhang, H.; Mukundan, S.; Mihaila, S.M.; Demarchi, D.; Khademhosseini, A. Highly elastomeric poly(glycerol sebacate)-co-poly(ethylene glycol) amphiphilic block copolymers. Biomaterials 2013, 34, 3970–3983. [Google Scholar] [CrossRef] [PubMed]
- Frydrych, M.; Román, S.; MacNeil, S.; Chen, B. Biomimetic poly(glycerol sebacate)/poly(l-lactic acid) blend scaffolds for adipose tissue engineering. Acta Biomater. 2015, 18, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Sundback, C.A.; Shyu, J.Y.; Wang, Y.; Faquin, W.C.; Langer, R.S.; Vacanti, J.P.; Hadlock, T.A. Biocompatibility analysis of poly(glycerol sebacate) as a nerve guide material. Biomaterials 2005, 26, 5454. [Google Scholar] [CrossRef] [PubMed]
- Redenti, S.; Neeley, W.L.; Rompani, S.; Saigal, S.; Yang, J.; Klassen, H.; Langer, R.; Young, M.J. Engineering retinal progenitor cell and scrollable poly(glycerol-sebacate) composites for expansion and subretinal transplantation. Biomaterials 2009, 30, 3405–3414. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-Z.; Bismarck, A.; Hansen, U.; Junaid, S.; Tran, M.Q.; Harding, S.E.; Ali, N.N.; Boccaccini, A.R. Characterisation of a soft elastomer poly(glycerol sebacate) designed to match the mechanical properties of myocardial tissue. Biomaterials 2008, 29, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, E.M.; Allen, R.A.; Gao, J.; Pesce, M.; Wang, Y. Highly elastic and suturable electrospun poly(glycerol sebacate) fibrous scaffolds. Acta Biomater. 2015, 18, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Elahi, F.; Lu, W.; Guoping, G.; Khan, F. Core-shell Fibers for Biomedical Applications—A Review. Bioeng. Biomed. Sci. J. 2013, 3, 1–14. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Chung, O.H.; Park, J.S. Coaxial electrospun poly(lactic acid)/chitosan (core/shell) composite nanofibers and their antibacterial activity. Carbohydr. Polym. 2011, 86, 1799–1806. [Google Scholar] [CrossRef]
- Xu, B.; Li, Y.; Fang, X.; Thouas, G.A.; Cook, W.D.; Newgreen, D.F.; Chen, Q. Mechanically tissue-like elastomeric polymers and their potential as a vehicle to deliver functional cardiomyocytes. J. Mech. Behav. Biomed. Mater. 2013, 28, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Griffin, M.A.; Sen, S.; Bönnemann, C.G.; Sweeney, H.L.; Discher, D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004, 166, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Wells, R.G. The role of matrix stiffness in regulating cell behavior. Hepatology 2008, 47, 1394–1400. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Hearing, V.J.; Itami, S.; Yoshikawa, K.; Katayama, I. Mesenchymal-epithelial interactions in the skin: Aiming for site-specific tissue regeneration. J. Dermatol. Sci. 2005, 40, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mao, J.J.; Chen, L. Epithelial-mesenchymal interactions as a working concept for oral mucosa regeneration. Tissue Eng. Part B. Rev. 2011, 17, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Jean-Gilles, R.; Soscia, D.; Sequeira, S.; Melfi, M.; Gadre, A.; Castracane, J.; Larsen, M. Novel Modeling Approach to Generate a Polymeric Nanofiber Scaffold for Salivary Gland Cells. J. Nanotechnol. Eng. Med. 2012, 1, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Sequeira, S.J.; Soscia, D.A.; Oztan, B.; Mosier, A.P.; Jean-Gilles, R.; Gadre, A.; Cady, N.C.; Yener, B.; Castracane, J.; Larsen, M. The regulation of focal adhesion complex formation and salivary gland epithelial cell organization by nanofibrous PLGA scaffolds. Biomaterials 2012, 33, 3175–3186. [Google Scholar] [CrossRef] [PubMed]
- Sfakis, L.; Kamaldinov, T.; Larsen, M.; Castracane, J.; Khmaladze, A. Quantification of Confocal images using LabVIEW for tissue engineering applications. Tissue Eng. Method C 2016, 11, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Sfakis, L.; Sharikova, A.; Tuschel, D.; Costa, F.X.; Larsen, M.; Khmaladze, A.; Castracane, J. Core/shell nanofiber characterization by Raman scanning microscopy. Biomed. Opt. Express 2017, 8, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Neelam, S.; Hayes, P.R.; Zhang, Q.; Dickinson, R.B.; Lele, T.P. Vertical uniformity of cells and nuclei in epithelial monolayers. Sci. Rep. 2016, 6, 19689. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ameer, G.A.; Sheppard, B.J.; Langer, R. A tough biodegradable elastomer. Nat. Biotechnol. 2002, 20, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Laoide, B.M.; Courty, Y.; Gastinne, I.; Thibaut, C.; Kellermann, O. Immortalised mouse submandibular epithelial cell lines retain polarised structural and functional properties. J. Cell Sci. 1996, 2800, 2789–2800. [Google Scholar]
- Todaro, G.J.; Green, H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963, 17, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lovett, D.; Zhang, Q.; Neelam, S.; Kuchibhotla, R.A.; Zhu, R.; Gundersen, G.G.; Lele, T.P.; Dickinson, R.B. Moving Cell Boundaries Drive Nuclear Shaping during Cell Spreading. Biophys. J. 2015, 109, 670–686. [Google Scholar] [CrossRef] [PubMed]







| Scaffold Type | Young’s Modulus (MPa) | Water Contact Angle (Mean ± Stdev) |
|---|---|---|
| PLGA | 4.8 ± 1.5 | 122 ± 4 |
| PGS/PLGA | 1.3 ± 0.66 | 65 ± 5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sfakis, L.; Kamaldinov, T.; Khmaladze, A.; Hosseini, Z.F.; Nelson, D.A.; Larsen, M.; Castracane, J. Mesenchymal Cells Affect Salivary Epithelial Cell Morphology on PGS/PLGA Core/Shell Nanofibers. Int. J. Mol. Sci. 2018, 19, 1031. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041031
Sfakis L, Kamaldinov T, Khmaladze A, Hosseini ZF, Nelson DA, Larsen M, Castracane J. Mesenchymal Cells Affect Salivary Epithelial Cell Morphology on PGS/PLGA Core/Shell Nanofibers. International Journal of Molecular Sciences. 2018; 19(4):1031. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041031
Chicago/Turabian StyleSfakis, Lauren, Tim Kamaldinov, Alexander Khmaladze, Zeinab F. Hosseini, Deirdre A. Nelson, Melinda Larsen, and James Castracane. 2018. "Mesenchymal Cells Affect Salivary Epithelial Cell Morphology on PGS/PLGA Core/Shell Nanofibers" International Journal of Molecular Sciences 19, no. 4: 1031. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041031