Exosomal miRNA Signatures for Late-Onset Acute Graft-Versus-Host Disease in Allogenic Hematopoietic Stem Cell Transplantation
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
2.2. miRNA Expression Profiling at LA GVHD Diagnosis
2.3. Comparison of Exosomal miRNAs Differentially Expressed among LA GVHD, Non-GVHD, and Healthy Volunteers
2.4. Quantification of Individual miRNA by Real-Time Quantitive RT-PCR
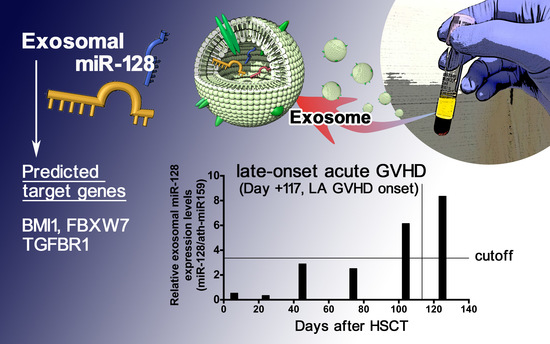
2.5. Sequential Analysis of Exosomal miRNAs Post-Allogenic HSCT
2.6. Prediction of the Target Genes of Candidate miRNAs
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Diagnosis of Late-Onset Acute GVHD
4.3. Isolation of Exosome Fractions from Plasma Sample of Patients
4.4. Exosomal miRNA Profiling
4.5. TaqMan Loq-Density Array Screening
4.6. Real-Time Quantitative RT-PCR for Candidate miRNAs
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Filipovich, A.; Weisdorf, D.; Pavletic, S.; Socie, G.; Wingard, J.R.; Lee, S.J.; Martin, P.; Chien, J.; Przepiorka, D.; Couriel, D.; et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol. Blood Marrow Transplant. 2005, 11, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Omer, A.K.; Weisdorf, D.J.; Lazaryan, A.; Shanley, R.; Blazar, B.R.; MacMillan, M.L.; Brunstein, C.; Bejanyan, N.; Arora, M. Late Acute Graft-versus-Host Disease after Allogeneic Hematopoietic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2016, 22, 879–883. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.S.; Lee, S.E.; Song, H.H.; Lee, J.H.; Yahng, S.A.; Eom, K.S.; Kim, Y.-J.; Kim, H.-J.; Lee, S.; Min, C.-K.; et al. Graft-versus-tumor according to type of graft-versus-host disease defined by National Institute of Health consensus criteria and associated outcome. Biol. Blood Marrow Transplant. 2012, 18, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Paczesny, S.; Krijanovski, O.I.; Braun, T.M.; Choi, S.W.; Clouthier, S.G.; Kuick, R.; Misek, D.E.; Cooke, K.R.; Kicko, C.L.; Weyand, A.; et al. A biomarker panel for acute graft-versus-host disease. Blood 2009, 113, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.R., Jr.; Khawaja, M.R.; Perkins, S.M.; Mumaw, C.L.; Orschell, C.; Paczesny, S. Prognostic biomarkers for acute graft-versus-host disease risk after cyclophosphamide-fludarabine nonmyeloablative allotransplantation. Biol Blood Marrow Transplant 2014, 20, 1861–1864. [Google Scholar] [CrossRef] [PubMed]
- Vader Lugt, M.T.; Braun, T.M.; Hanash, S.; Ritz, J.; Ho, V.T.; Antin, J.H.; Zhang, Q.; Wong, C.-H.; Wang, H.; Chin, A.; et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. N. Engl. J. Med. 2013, 369, 529–539. [Google Scholar] [CrossRef] [PubMed]
- McDonald, G.B.; Tabellini, L.; Storer, B.E.; Lawler, R.L.; Martin, P.J.; Hansen, J.A. Plasma biomarkers of acute GVHD and nonrelapse mortality: Predictive value of measurements before GVHD onset and treatment. Blood 2015, 126, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosome: Extracellular organelles important in intracellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Farber, E.L.; Rapoport, A.L.; Tejada, D.; Deniskin, R.; Akhmedov, N.B.; Farber, D.B. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS ONE 2009, 4, e4722. [Google Scholar] [CrossRef] [PubMed]
- Umezu, T.; Tadokoro, H.; Azuma, K.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014, 124, 3748–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAa and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666–80678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sueta, A.; Yamamoto, Y.; Tomiguchi, M.; Takeshita, T.; Yamamoto-Ibusuki, M.; Iwase, H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget 2017, 8, 69934–69944. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.; Heaphy, C.E.; Costinean, S.; Stauffer, N.; Na, C.; Hamadani, M.; Santhanam, R.; Mao, C.; Taylor, P.A.; Sandhu, S.; et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood 2012, 119, 4786–4797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Robinson, S.N.; Setoyama, T.; Tung, S.S.; D’Abundo, L.; Shah, M.Y.; Yang, H.; Yvon, E.; Shah, N.; Yang, H.; et al. FOXP3 is a direct target of miR15a/16 in umbilical cord blood regulatory T cells. Bone Marrow Transplant 2014, 49, 793–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Heinrichs, J.; Bastian, D.; Fu, J.; Nguyen, H.; Schutt, S.; Liu, Y.; Jin, J.; Liu, C.; Li, Q.-J.; et al. MicroRNA-17-92 controls T-cell responses in graft-versus-host disease and leukemia relapse in mice. Blood 2015, 126, 1314–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, B.; Wang, Y.; Li, W.; Baker, M.; Guo, J.; Corbet, K.; Tsalik, E.L.; Li, Q.-J.; Palmer, S.M.; Wood, C.W.; et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood 2013, 122, 3365–3375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhao, X.; Ye, X.; Luo, H.; Zhao, T.; Diao, Y.; Zhang, H.; Lv, M.; Zhang, W.; Huang, X.; et al. Plasma microRNA-586 is a new biomarker for acute graft-versus-host disease. Ann. Hematol. 2015, 94, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Crossland, R.E.; Norden, J.; Juric, M.K.; Green, K.; Pearce, K.F.; Lendrem, C.; Greinix, H.T.; Dickinson, A.M. Expression of Serum microRNAs is Altered During Acute Graft-versus-Host Disease. Front. Immunol. 2017, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Oravecz-Wilson, K.; Mathewson, N.; Wang, Y.; McEachin, R.; Liu, C.; Toubai, T.; Wu, J.; Rossi, C.; Braun, T.; et al. Mature T cell responses are controlled by microRNA-142. J. Clin. Investig. 2015, 125, 2825–2840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Williams, S.; Otsuki, A.; Nuovo, G.; Raychaudhury, A.; Newton, H.B.; Chiocca, E.A.; Lawler, S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008, 68, 9125–9130. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.G.; Zhao, Y.; Sethi, P.; Li, Y.Y.; Mahta, A.; Culicchia, F.; Lukiw, W.J. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J. Neurooncol. 2010, 98, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Zhang, T.; Liu, C.; Badeaux, M.A.; Liu, B.; Liu, R.F.; Jeter, C.; Chen, X.; Vlassov, A.V.; Tang, D.G. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014, 74, 4183–4195. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; He, X.; Zou, J.; Guo, P.; Jiang, S.; Lv, N.; Alexseyev, Y.; Luo, L.; Luo, Z. Negative regulation of Bmi-1 by AMPK and implication in cancer progression. Oncotarget 2016, 7, 6188–6200. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Wilfred, B.R.; Hu, Y.; Stromberg, A.J.; Nelson, P.T. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA 2010, 16, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.-C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Lia, G.; Brunello, L.; Bruno, S.; Carpanetto, A.; Omede, P.; Festuccia, M.; Tosti, L.; Maffini, E.; Giaccone, L.; Arpinati, M.; et al. Extracellular vesicles as potentiall biomarkers of acute graft-vs-host disease. Leukemia 2018, 32, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, K.; Umezu, T.; Katagiri, S.; Kobayashi, C.; Azuma, K.; Tauchi, T.; Okabe, S.; Fukuoka, Y.; Oyashika, J.H. Down-regulation of plasma miR-125 in chronic myeloid leukemia patients with successful discontinuation of imatinib. Int. J. Mol. Sci. 2016, 17, 570. [Google Scholar] [CrossRef] [PubMed]
- Kordelas, L.; Rebmann, V.; Ludwig, A.K.; Radtke, S.; Ruesing, J.; Doeppner, T.R.; Epple, M.; Horn, P.A.; Beelen, D.W.; Giebel, B. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 2014, 28, 970–973. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, J.H.; Ohtsuki, K.; Mizoguchi, I.; Yoshimoto, T.; Katagiri, S.; Umezu, T.; Ohyashiki, K. Downregulated microRNA-148b in circulating PBMCs in chronic myeloid leukemia patients with undetectable minimal residual disease: A possible biomarker to discontinue imatinib safely. Drug Des. Dev. Ther. 2014, 8, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, S.; Ohyashiki, J.H.; Ohyashiki, M.; Umezu, T.; Suzuki, K.; Inagaki, A.; Lida, S.; Ohyashiki, K. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J. 2012, 2, e53. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, K.; Umezu, T.; Yoshizawa, S.; Ito, Y.; Ohyashiki, M.; Kawashima, H.; Tanaka, M.; Kuroda, M.; Ohyashiki, J.H. Clinical impact of down-regulated plasma miR-92a levels in non-Hodgkin’s lymphoma. PLoS ONE 2011, 6, e16408. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Gräfe, A.; Seiler, A.; Schumacher, S.; Nitsch, R.; Wulczyn, F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005, 21, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.P.; Poisson, L.M.; Bhat, V.B.; Fermin, D.; Zhao, R.; Kalyana-Sundaram, S.; Michailidis, G.; Nesvizhskii, A.I.; Omenn, G.S.; Chinnaiyan, A.M.; et al. Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol. Cell Proteom. 2010, 9, 298–312. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.H.; László, C.F.; Greco, S.; Chambers, S.K. Regulation of colony stimulating factor-1 expression and ovarian cancer cell behavior in vitro by miR-128 and miR-152. Mol. Cancer 2012, 11, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, P.; Wischhsen, J.; Happold, C.; Chandran, P.A.; Hofer, S.; Eisele, G.; Weller, M.; Keller, A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J. Neurochem. 2011, 118, 449–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razo, A.; Dontje, B.; Vellenga, E.; de Haan, G.; Schuringa, J.J. Long-term maintenance of human hematopoietic stem/progenitor cells by expression of BMI1. Blood 2008, 111, 2621–2630. [Google Scholar] [CrossRef] [PubMed]







| Diagnosis | Age (y) | Gender | Donor | HLA Sero Compatibility | Stem Cell Source | Conditioning | GVHD Prophylaxis | Days of LA GVHD Onset | Days of Sampling | Target Organ | Grade | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Late-Onset Acute GVHD Group | ||||||||||||
| PN 1 | AML | 33 | M | Matched unrelated | 6/6 | BM | Myeloablative | CsA + st-MTX | +238 | +248 | Liver | IV |
| PN 2 | NHL | 38 | M | Matched related | 6/6 | PBSC | Reduced intensity | CsA + st-MTX | +180 | +221 | Liver | IV |
| PN 3 | AML | 57 | M | Mismatched | 5/6 | BM | Myeloablative | Tac + st-MTX | +117 | +136 | Gut | III |
| PN 4 | ALL | 56 | F | Mismatched | 4/6 | Umbilical cord blood | Myeloablative | Tac | +119 | +132 | Skin + gut | IV |
| PN 5 | ALL | 60 | F | Mismatched | 4/6 | Umbilical cord blood | Myeloablative | Tac | +165 | +174 | gut | II |
| Non-GVHD Group | ||||||||||||
| PN 6 | ALL | 62 | M | Matched unrelated | 6/6 | BM | Reduced intensity | CsA + st-MTX | +42 | |||
| PN 7 | ALL | 26 | F | Matched unrelated | 6/6 | BM | Myeloablative | CsA + st-MTX | +41 | |||
| PN 8 | AML | 60 | M | Mismatched | 4/6 | Umbilical cord blood | Myeloablative | Tac | +49 | |||
| PN 9 | AML | 62 | M | Mismatched | 5/6 | PBSC | Reduced intensity | Tac + st-MTX | +41 | |||
| PN 10 | AML | 70 | F | Mismatched | 4/6 | Umbilical cord blood | Reduced intensity | Tac | +45 | |||
| Exosomal miRNAs | Fold Change | p Value |
|---|---|---|
| hsa-miR-423-5p | 49.07 | 0.000198 |
| hsa-miR-19a | 37.33 | 0.000013 |
| hsa-miR-142-3p | 28.35 | 0.000198 |
| hsa-miR-128 | 24.79 | 0.000111 |
| hsa-miR-193b | 16.12 | 0.000209 |
| hsa-miR-30c | 14.33 | 0.003157 |
| hsa-miR-193a | 12.56 | 0.000111 |
| hsa-miR-191 | 12.3 | 0.000081 |
| hsa-miR-125b | 10.7 | 0.001064 |
| hsa-miR-574-3p | 6.13 | 0.000198 |
| miRNA | Experimentally Validated Targets by Reporter Assay (miRTarBase) |
|---|---|
| hsa-miR-128 | BMI1, FBXW7, DCX, RELN, WEE1, TGFBR1, NEK2, SREBF1, SREBF2, ABCA1, ABCG1, RXRA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshizawa, S.; Umezu, T.; Saitoh, Y.; Gotoh, M.; Akahane, D.; Kobayashi, C.; Ohyashiki, J.H.; Ohyashiki, K. Exosomal miRNA Signatures for Late-Onset Acute Graft-Versus-Host Disease in Allogenic Hematopoietic Stem Cell Transplantation. Int. J. Mol. Sci. 2018, 19, 2493. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092493
Yoshizawa S, Umezu T, Saitoh Y, Gotoh M, Akahane D, Kobayashi C, Ohyashiki JH, Ohyashiki K. Exosomal miRNA Signatures for Late-Onset Acute Graft-Versus-Host Disease in Allogenic Hematopoietic Stem Cell Transplantation. International Journal of Molecular Sciences. 2018; 19(9):2493. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092493
Chicago/Turabian StyleYoshizawa, Seiichiro, Tomohiro Umezu, Yuu Saitoh, Moritaka Gotoh, Daigo Akahane, Chiaki Kobayashi, Junko H. Ohyashiki, and Kazuma Ohyashiki. 2018. "Exosomal miRNA Signatures for Late-Onset Acute Graft-Versus-Host Disease in Allogenic Hematopoietic Stem Cell Transplantation" International Journal of Molecular Sciences 19, no. 9: 2493. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092493