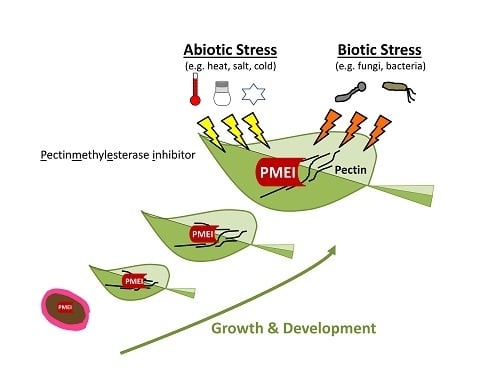
The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs)
Abstract
:1. Introduction
2. PMEI Occurrence and Regulation
3. PME Inhibitor Structure and Interaction with Pectin Esterases
4. Role of PMEIs in Stress
5. Role of PMEIs in Development
5.1. PMEI Function in Seeds
5.2. PMEI Function in Growth Processes
5.3. PMEI Function in Organ Formation
5.4. PMEIs in Fruit Development
6. Potential of PMEIs for Applications
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PMEI | Pectin methylesterase inhibitor |
| PME | Pectin methyl esterase |
| DM | Degree of methylation |
| HG | Homogalacturonan |
| GalA | D-galacturonic acid |
| GAUT | Galacturonosyltransferases |
| OGs | Oligogalacturonides |
| DAMP | Damage associated molecular pattern |
| H2O2 | Hydrogen peroxide |
| SCE | Seed coat epidermis |
| MUM1 | Mucilage-modified mutant 1 |
References
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Braybrook, S.A.; Hofte, H.; Peaucelle, A. Probing the mechanical contributions of the pectin matrix. Plant Signal. Behav. 2012, 7, 1037–1041. [Google Scholar] [CrossRef]
- Palin, R.; Geitmann, A. The role of pectin in plant morphogenesis. Biosystems 2012, 109, 397–402. [Google Scholar] [CrossRef]
- Daher, F.B.; Braybrook, S.A. How to let go: Pectin and plant cell adhesion. Front. Plant Sci. 2015, 6, 523. [Google Scholar] [CrossRef]
- Levesque-Tremblay, G.; Pelloux, J.; Braybrook, S.A.; Müller, K. Tuning of pectin methylesterification: Consequences for cell wall biomechanics and development. Planta 2015, 242, 791–811. [Google Scholar] [CrossRef]
- Saffer, A.M. Expanding roles for pectins in plant development. Integr. Plant Biol. 2018. [Google Scholar] [CrossRef]
- Sterling, J.D.; Quigley, H.F.; Orellana, A.; Mohnen, D. The catalytic site of the pectin biosynthetic enzyme alpha-1,4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol. 2001, 127, 360–371. [Google Scholar]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kumar Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Sakuragi, Y.; Zhu, X.; Burrell, A.J.; Mohanty, S.S.; Atwood, J.A.; Orlando, R.; Scheller, H.V.; Mohnen, D. Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc. Natl. Acad. Sci. USA 2011, 108, 20225–20230. [Google Scholar] [CrossRef]
- Biswal, A.K.; Atmodjo, M.A.; Li, M.; Baxter, H.L.; Yoo, C.G.; Pu, Y.; Lee, Y.-C.; Mazarei, M.; Black, I.M.; Zhang, J.-Y.; et al. Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat. Biotechnol. 2018, 36, 249–257. [Google Scholar] [CrossRef]
- Mouille, G.; Ralet, M.-C.; Cavelier, C.; Eland, C.; Effroy, D.; Hématy, K.; McCartney, L.; Truong, H.N.; Gaudon, V.; Thibault, J.-F.; et al. Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 2007, 50, 605–614. [Google Scholar] [CrossRef]
- Krupková, E.; Immerzeel, P.; Pauly, M.; Schmülling, T. The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J. 2007, 50, 735–750. [Google Scholar] [CrossRef]
- Ralet, M.-C.; Crépeau, M.-J.; Buchholt, H.-C.; Thibault, J.-F. Polyelectrolyte behaviour and calcium binding properties of sugar beet pectins differing in their degrees of methylation and acetylation. Biochem. Eng. J. 2003, 16, 191–201. [Google Scholar] [CrossRef]
- De Souza, A.J.; Pauly, M. Comparative genomics of pectinacetylesterases: Insight on function and biology. Plant Signal. Behav. 2015, 10, e1055434. [Google Scholar] [CrossRef]
- Gou, J.-Y.; Miller, L.M.; Hou, G.; Yu, X.-H.; Chen, X.-Y.; Liu, C.-J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef]
- Braccini, I.; Pérez, S. Molecular basis of Ca2+-induced gelation in alginates and pectins: The egg-box model revisited. Biomacromolecules 2001, 2, 1089–1096. [Google Scholar] [CrossRef]
- White, P.B.; Wang, T.; Park, Y.B.; Cosgrove, D.J.; Hong, M. Water–polysaccharide interactions in the primary cell wall of Arabidopsis thaliana from polarization transfer solid-state NMR. J. Am. Chem. Soc. 2014, 136, 10399–10409. [Google Scholar] [CrossRef]
- Ha, M.-A.; Apperley, D.C.; Jarvis, M.C. Molecular rigidity in dry and hydrated onion cell walls. Plant Physiol. 1997, 115, 593–598. [Google Scholar]
- Tibbits, C.W.; MacDougall, A.J.; Ring, S.G. Calcium binding and swelling behaviour of a high methoxyl pectin gel. Carbohydr. Res. 1998, 310, 101–107. [Google Scholar] [CrossRef]
- Peaucelle, A.; Braybrook, S.A.; Le Guillou, L.; Bron, E.; Kuhlemeier, C.; Höfte, H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef]
- Amsbury, S.; Hunt, L.; Elhaddad, N.; Baillie, A.; Lundgren, M.; Verhertbruggen, Y.; Scheller, H.V.; Knox, J.P.; Fleming, A.J.; Gray, J.E. Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Curr. Biol. 2016, 26, 2899–2906. [Google Scholar] [CrossRef]
- Carter, R.; Woolfenden, H.; Baillie, A.; Amsbury, S.; Carroll, S.; Healicon, E.; Sovatzoglou, S.; Braybrook, S.; Gray, J.E.; Hobbs, J.; et al. Stomatal opening involves polar, not radial, stiffening of guard cells. Curr. Biol. 2017, 27, 2974.e2–2983.e2. [Google Scholar] [CrossRef]
- Peaucelle, A.; Louvet, R.; Johansen, J.N.; Höfte, H.; Laufs, P.; Pelloux, J.; Mouille, G. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 2008, 18, 1943–1948. [Google Scholar] [CrossRef]
- Peaucelle, A.; Wightman, R.; Höfte, H. The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr. Biol. 2015, 25, 1746–1752. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Camardella, L.; Giovane, A.; Obel, N.; Pauly, M.; Favaron, F.; Cervone, F.; Bellincampi, D. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 2007, 143, 1871–1880. [Google Scholar] [CrossRef]
- Osorio, S.; Castillejo, C.; Quesada, M.A.; Medina-Escobar, N.; Brownsey, G.J.; Suau, R.; Heredia, A.; Botella, M.A.; Valpuesta, V. Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). Plant J. 2008, 54, 43–55. [Google Scholar] [CrossRef]
- Sénéchal, F.; Wattier, C.; Rustérucci, C.; Pelloux, J. Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Dean, G.H.; Griffiths, J.S.; Kirchsteiger, K.; Hwang, Y.T.; Gillett, A.; Dow, G.; Western, T.L.; Estelle, M.; Haughn, G.W. FLYING SAUCER1 Is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 2013, 25, 944–959. [Google Scholar] [CrossRef]
- Sénéchal, F.; L’Enfant, M.; Domon, J.-M.; Rosiau, E.; Crépeau, M.-J.; Surcouf, O.; Esquivel-Rodriguez, J.; Marcelo, P.; Mareck, A.; Guérineau, F.; et al. Tuning of pectin methylesterification. J. Biol. Chem. 2015, 290, 23320–23335. [Google Scholar] [CrossRef]
- Juge, N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 2006, 11, 359–367. [Google Scholar] [CrossRef]
- Pelloux, J.; Rustérucci, C.; Mellerowicz, E.J. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef]
- Balestrieri, C.; Castaldo, D.; Giovane, A.; Quagliuolo, L.; Servillo, L. A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur. J. Biochem. 1990, 193, 183–187. [Google Scholar]
- Wang, M.; Yuan, D.; Gao, W.; Li, Y.; Tan, J.; Zhang, X. A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS ONE 2013, 8, e72082. [Google Scholar] [CrossRef]
- Liu, T.; Yu, H.; Xiong, X.; Yue, X.; Yu, Y.; Huang, L.; Cao, J. Genome-wide identification, molecular evolution, and expression profiling analysis of pectin methylesterase inhibitor genes in Brassica campestris ssp. chinensis. Int. J. Mol. Sci. 2018, 19, 1338. [Google Scholar] [CrossRef]
- Tan, C.; Liu, Z.; Huang, S.; Li, C.; Ren, J.; Tang, X.; Liu, W.; Peng, S.; Feng, H. Pectin methylesterase inhibitor (PMEI) family can be related to male sterility in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. 2018, 293, 343–357. [Google Scholar] [CrossRef]
- Pinzón-Latorre, D.; Deyholos, M.K. Characterization and transcript profiling of the pectin methylesterase (PME) and pectin methylesterase inhibitor (PMEI) gene families in flax (Linum usitatissimum). BMC Genom. 2013, 14, 742. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Jeong, H.Y.; Kim, H.; Kim, Y.C.; Lee, C. Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs). Plant Physiol. Biochem. 2016, 101, 105–112. [Google Scholar] [CrossRef]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Mattei, B.; Bellincampi, D. The grapevine VvPMEI1 gene encodes a novel functional pectin methylesterase inhibitor associated to grape berry development. PLoS ONE 2015, 10, e0133810. [Google Scholar] [CrossRef]
- Rocchi, V.; Janni, M.; Bellincampi, D.; Giardina, T.; D’Ovidio, R. Intron retention regulates the expression of pectin methyl esterase inhibitor (PMEI) genes during wheat growth and development. Plant Biol. 2012, 14, 365–373. [Google Scholar] [CrossRef]
- Pinzon-Latorre, D.; Deyholos, M.K. Pectinmethylesterases (PME) and pectinmethylesterase inhibitors (PMEI) enriched during phloem fiber development in flax (Linum usitatissimum). PLoS ONE 2014, 9, e105386. [Google Scholar] [CrossRef]
- Saez-Aguayo, S.; Ralet, M.-C.; Berger, A.; Botran, L.; Ropartz, D.; Marion-Poll, A.; North, H.M. PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 2013, 25, 308–323. [Google Scholar] [CrossRef]
- Shi, D.; Ren, A.; Tang, X.; Qi, G.; Xu, Z.; Chai, G.; Hu, R.; Zhou, G.; Kong, Y. MYB52 negatively regulates pectin demethylesterification in seed coat mucilage. Plant Physiol. 2018, 176, 2737–2749. [Google Scholar] [CrossRef]
- Srivastava, S.; Gupta, S.M.; Sane, A.P.; Nath, P. Isolation and characterization of ripening related pectin methylesterase inhibitor gene from banana fruit. Physiol. Mol. Biol. Plants 2012, 18, 191–195. [Google Scholar] [CrossRef] [Green Version]
- Lionetti, V.; Fabri, E.; De Caroli, M.; Hansen, A.R.; Willats, W.G.T.; Piro, G.; Bellincampi, D. Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol. 2017, 173, 1844–1863. [Google Scholar] [CrossRef]
- Hong, M.J.; Kim, D.Y.; Lee, T.G.; Jeon, W.B.; Seo, Y.W. Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat. Genes Genet. Syst. 2010, 85, 97–106. [Google Scholar] [Green Version]
- An, S.H.; Choi, H.W.; Hong, J.K.; Hwang, B.K. Regulation and function of the pepper pectin methylesterase inhibitor (CaPMEI1) gene promoter in defense and ethylene and methyl jasmonate signaling in plants. Planta 2009, 230, 1223–1237. [Google Scholar] [CrossRef]
- Bosch, M.; Hepler, P.K. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 2005, 17, 3219–3226. [Google Scholar] [CrossRef]
- Parre, E.; Geitmann, A. Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta 2005, 220, 582–592. [Google Scholar] [CrossRef]
- Röckel, N.; Wolf, S.; Kost, B.; Rausch, T.; Greiner, S. Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 2008, 53, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Hothorn, M.; Wolf, S.; Aloy, P.; Greiner, S.; Scheffzek, K. Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 2004, 16, 3437–3447. [Google Scholar] [CrossRef]
- Giovane, A.; Servillo, L.; Balestrieri, C.; Raiola, A.; D’Avino, R.; Tamburrini, M.; Ciardiello, M.A.; Camardella, L. Pectin methylesterase inhibitor. Biochim. Biophys. Acta 2004, 1696, 245–252. [Google Scholar] [CrossRef]
- Camardella, L.; Carratore, V.; Ciardiello, M.A.; Servillo, L.; Balestrieri, C.; Giovane, A. Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur. J. Biochem. 2000, 267, 4561–4565. [Google Scholar]
- Di Matteo, A.; Giovane, A.; Raiola, A.; Camardella, L.; Bonivento, D.; De Lorenzo, G.; Cervone, F.; Bellincampi, D.; Tsernoglou, D. Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 2005, 17, 849–858. [Google Scholar] [CrossRef]
- Raiola, A.; Camardella, L.; Giovane, A.; Mattei, B.; De Lorenzo, G.; Cervone, F.; Bellincampi, D. Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett. 2004, 557, 199–203. [Google Scholar]
- Jolie, R.P.; Duvetter, T.; Van Loey, A.M.; Hendrickx, M.E. Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 2010, 345, 2583–2595. [Google Scholar] [CrossRef]
- Reca, I.B.; Lionetti, V.; Camardella, L.; D’Avino, R.; Giardina, T.; Cervone, F.; Bellincampi, D. A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1. Plant Mol. Biol. 2012, 79, 429–442. [Google Scholar] [CrossRef]
- Bonavita, A.; Carratore, V.; Ciardiello, M.A.; Giovane, A.; Servillo, L.; D’Avino, R. Influence of pH on the structure and function of kiwi pectin methylesterase inhibitor. J. Agric. Food Chem. 2016, 64, 5866–5876. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Jeong, H.Y.; Jeon, S.H.; Kim, D.; Lee, C. Rice pectin methylesterase inhibitor28 (OsPMEI28) encodes a functional PMEI and its overexpression results in a dwarf phenotype through increased pectin methylesterification levels. J. Plant Physiol. 2017, 208, 17–25. [Google Scholar] [CrossRef]
- Sénéchal, F.; Mareck, A.; Marcelo, P.; Lerouge, P.; Pelloux, J. Arabidopsis PME17 activity can be controlled by pectin methylesterase inhibitor 4. Plant Signal. Behav. 2015, 10, e983351. [Google Scholar] [CrossRef]
- Hocq, L.; Sénéchal, F.; Lefebvre, V.; Lehner, A.; Domon, J.-M.; Mollet, J.-C.; Dehors, J.; Pageau, K.; Marcelo, P.; Guérineau, F.; et al. Combined experimental and computational approaches reveal distinct pH dependence of pectin methylesterase inhibitors. Plant Physiol. 2017, 173, 1075–1093. [Google Scholar] [CrossRef]
- Hamann, T. The plant cell wall integrity maintenance mechanism—A case study of a cell wall plasma membrane signaling network. Phytochemistry 2015, 112, 100–109. [Google Scholar] [CrossRef]
- Espino, J.J.; Gutiérrez-Sánchez, G.; Brito, N.; Shah, P.; Orlando, R.; González, C. The Botrytis cinerea early secretome. Proteomics 2010, 10, 3020–3034. [Google Scholar] [CrossRef]
- D’Ovidio, R.; Mattei, B.; Roberti, S.; Bellincampi, D. Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant–pathogen interactions. Biochim. Biophys. Acta 2004, 1696, 237–244. [Google Scholar] [CrossRef]
- Lionetti, V. PECTOPLATE: The simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Front. Plant Sci. 2015, 6, 331. [Google Scholar] [CrossRef]
- Raiola, A.; Lionetti, V.; Elmaghraby, I.; Immerzeel, P.; Mellerowicz, E.J.; Salvi, G.; Cervone, F.; Bellincampi, D. Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol. Plant Microbe Interact. 2011, 24, 432–440. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, X.; Sun, Y.; Wang, P.; Li, X.; Pei, Y.; Li, F.; Hou, Y. Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 2017, 7, 39840. [Google Scholar] [CrossRef]
- Liu, C.-Q.; Hu, K.-D.; Li, T.-T.; Yang, Y.; Yang, F.; Li, Y.-H.; Liu, H.-P.; Chen, X.-Y.; Zhang, H. Polygalacturonase gene pgxB in Aspergillus niger is a virulence factor in apple fruit. PLoS ONE 2017, 12, e0173277. [Google Scholar] [CrossRef]
- Bravo Ruiz, G.; Di Pietro, A.; Roncero, M.I.G. Combined action of the major secreted exo- and endopolygalacturonases is required for full virulence of Fusarium oxysporum. Mol. Plant Pathol. 2016, 17, 339–353. [Google Scholar] [CrossRef]
- Cho, Y.; Jang, M.; Srivastava, A.; Jang, J.-H.; Soung, N.-K.; Ko, S.-K.; Kang, D.-O.; Ahn, J.S.; Kim, B.Y. A pectate lyase-coding gene abundantly expressed during early stages of infection is required for full virulence in Alternaria brassicicola. PLoS ONE 2015, 10, e0127140. [Google Scholar] [CrossRef]
- Bethke, G.; Grundman, R.E.; Sreekanta, S.; Truman, W.; Katagiri, F.; Glazebrook, J. Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol. 2014, 164, 1093–1107. [Google Scholar] [CrossRef]
- Ferrari, S.; Savatin, D.V.; Sicilia, F.; Gramegna, G.; Cervone, F.; De Lorenzo, G. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013, 4, 49. [Google Scholar] [CrossRef]
- Kohorn, B.D.; Kohorn, S.L.; Saba, N.J.; Martinez, V.M. Requirement for pectin methyl esterase and preference for fragmented over native pectins for wall-associated kinase-activated, EDS1/PAD4-dependent stress response in Arabidopsis. J. Biol. Chem. 2014, 289, 18978–18986. [Google Scholar] [CrossRef]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [Green Version]
- Osorio, S.; Bombarely, A.; Giavalisco, P.; Usadel, B.; Stephens, C.; Aragüez, I.; Medina-Escobar, N.; Botella, M.A.; Fernie, A.R.; Valpuesta, V. Demethylation of oligogalacturonides by FaPE1 in the fruits of the wild strawberry Fragaria vesca triggers metabolic and transcriptional changes associated with defence and development of the fruit. J. Exp. Bot. 2011, 62, 2855–2873. [Google Scholar] [CrossRef]
- Komarova, T.V.; Sheshukova, E.V.; Dorokhov, Y.L. Cell wall methanol as a signal in plant immunity. Front. Plant Sci. 2014, 5, 101. [Google Scholar] [CrossRef]
- Volpi, C.; Janni, M.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant Microbe Interact. 2011, 24, 1012–1019. [Google Scholar] [CrossRef]
- Tundo, S.; Kalunke, R.; Janni, M.; Volpi, C.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. Pyramiding PvPGIP2 and TAXI-III but not PvPGIP2 and PMEI enhances resistance against Fusarium graminearum. Mol. Plant-Microbe Interact. 2016, 29, 629–639. [Google Scholar] [CrossRef]
- Marzin, S.; Hanemann, A.; Sharma, S.; Hensel, G.; Kumlehn, J.; Schweizer, G.; Röder, M.S. Are PECTIN ESTERASE INHIBITOR genes involved in mediating resistance to Rhynchosporium commune in barley? PLoS ONE 2016, 11, e0150485. [Google Scholar] [CrossRef]
- Lionetti, V.; Cervone, F.; Bellincampi, D. Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 2012, 169, 1623–1630. [Google Scholar] [CrossRef]
- An, S.H.; Sohn, K.H.; Choi, H.W.; Hwang, I.S.; Lee, S.C.; Hwang, B.K. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 2008, 228, 61–78. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-H.; Citovsky, V. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 2003, 35, 386–392. [Google Scholar] [Green Version]
- Lionetti, V.; Raiola, A.; Cervone, F.; Bellincampi, D. Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol. Plant Pathol. 2014, 15, 265–274. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014, 5, 771. [Google Scholar] [CrossRef]
- Jithesh, M.N.; Wally, O.S.D.; Manfield, I.; Critchley, A.T.; Hiltz, D.; Prithiviraj, B. Analysis of seaweed extract-induced transcriptome leads to identification of a negative regulator of salt tolerance in Arabidopsis. HortScience 2012, 47, 704–709. [Google Scholar]
- Chen, J.; Chen, X.; Zhang, Q.; Zhang, Y.; Ou, X.; An, L.; Feng, H.; Zhao, Z. A cold-induced pectin methyl-esterase inhibitor gene contributes negatively to freezing tolerance but positively to salt tolerance in Arabidopsis. J. Plant Physiol. 2018, 222, 67–78. [Google Scholar] [CrossRef]
- Solecka, D.; Zebrowski, J.; Kacperska, A. Are pectins involved in cold acclimation and de-acclimation of winter oil-seed rape plants? Ann. Bot. 2008, 101, 521–530. [Google Scholar] [CrossRef]
- Baldwin, L.; Domon, J.-M.; Klimek, J.F.; Fournet, F.; Sellier, H.; Gillet, F.; Pelloux, J.; Lejeune-Hénaut, I.; Carpita, N.C.; Rayon, C. Structural alteration of cell wall pectins accompanies pea development in response to cold. Phytochemistry 2014, 104, 37–47. [Google Scholar] [CrossRef]
- Qu, T.; Liu, R.; Wang, W.; An, L.; Chen, T.; Liu, G.; Zhao, Z. Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress. Cryobiology 2011, 63, 111–117. [Google Scholar] [CrossRef]
- North, H.M.; Berger, A.; Saez-Aguayo, S.; Ralet, M.-C. Understanding polysaccharide production and properties using seed coat mutants: Future perspectives for the exploitation of natural variants. Ann. Bot. 2014, 114, 1251–1263. [Google Scholar] [CrossRef]
- Western, T.L.; Skinner, D.J.; Haughn, G.W. Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 2000, 122, 345–356. [Google Scholar]
- Francoz, E.; Ranocha, P.; Burlat, V.; Dunand, C. Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci. 2015, 20, 515–524. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Yang, B.; Schmidt, M.H.-W.; Günl, M.; Usadel, B. Starting to gel: How Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int. J. Mol. Sci. 2015, 16, 3452–3473. [Google Scholar] [CrossRef] [Green Version]
- Turbant, A.; Fournet, F.; Lequart, M.; Zabijak, L.; Pageau, K.; Bouton, S.; Van Wuytswinkel, O. PME58 plays a role in pectin distribution during seed coat mucilage extrusion through homogalacturonan modification. J. Exp. Bot. 2016, 67, 2177–2190. [Google Scholar] [CrossRef] [Green Version]
- Rautengarten, C.; Usadel, B.; Neumetzler, L.; Hartmann, J.; Büssis, D.; Altmann, T. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 2008, 54, 466–480. [Google Scholar] [CrossRef]
- Müller, K.; Levesque-Tremblay, G.; Bartels, S.; Weitbrecht, K.; Wormit, A.; Usadel, B.; Haughn, G.; Kermode, A.R. Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiol. 2013, 161, 305–316. [Google Scholar] [CrossRef]
- Scheler, C.; Weitbrecht, K.; Pearce, S.P.; Hampstead, A.; Büttner-Mainik, A.; Lee, K.J.D.; Voegele, A.; Oracz, K.; Dekkers, B.J.W.; Wang, X.; et al. Promotion of testa rupture during garden cress germination involves seed compartment-specific expression and activity of pectin methylesterases. Plant Physiol. 2015, 167, 200–215. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Pauly, M.; Usadel, B. Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol. 2018, 176, 2590–2600. [Google Scholar] [CrossRef]
- Wolf, S.; Greiner, S. Growth control by cell wall pectins. Protoplasma 2012, 249, 169–175. [Google Scholar] [CrossRef]
- Pelletier, S.; van Orden, J.; Wolf, S.; Vissenberg, K.; Ndong, Y.A.; Pelloux, J.; Bischoff, V.; Urbain, A.; Lemonnier, G.; Renou, J.; et al. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 2010, 188, 726–739. [Google Scholar] [CrossRef] [Green Version]
- Paynel, F.; Leroux, C.; Surcouf, O.; Schaumann, A.; Pelloux, J.; Driouich, A.; Mollet, J.C.; Lerouge, P.; Lehner, A.; Mareck, A. Kiwi fruit PMEI inhibits PME activity, modulates root elongation and induces pollen tube burst in Arabidopsis thaliana. Plant Growth Regul. 2014, 74, 285–297. [Google Scholar] [CrossRef]
- Lionetti, V.; Francocci, F.; Ferrari, S.; Volpi, C.; Bellincampi, D.; Galletti, R.; D’Ovidio, R.; De Lorenzo, G.; Cervone, F. Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Natl. Acad. Sci. USA 2010, 107, 616–621. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Yang, S.-L.; Xie, L.-F.; Puah, C.S.; Zhang, X.-Q.; Yang, W.-C.; Sundaresan, V.; Ye, D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 2005, 17, 584–596. [Google Scholar] [CrossRef]
- Tian, G.-W.; Chen, M.-H.; Zaltsman, A.; Citovsky, V. Pollen-specific pectin methylesterase involved in pollen tube growth. Dev. Biol. 2006, 294, 83–91. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Feng, J.; Wu, J.; Wang, X.W. BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen tube growth. Planta 2010, 231, 1323–1334. [Google Scholar] [CrossRef]
- Chebli, Y.; Geitmann, A. Cellular growth in plants requires regulation of cell wall biochemistry. Curr. Opin. Cell Biol. 2017, 44, 28–35. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, W.; Traas, J. Mechanical signaling in plant morphogenesis. Curr. Opin. Genet. Dev. 2018, 51, 26–30. [Google Scholar] [CrossRef]
- Müller, K.; Levesque-Tremblay, G.; Fernandes, A.; Wormit, A.; Bartels, S.; Usadel, B.; Kermode, A. Overexpression of a pectin methylesterase inhibitor in Arabidopsis thaliana leads to altered growth morphology of the stem and defective organ separation. Plant Signal. Behav. 2013, 8, e26464. [Google Scholar] [CrossRef]
- Wolf, S.; Mravec, J.; Greiner, S.; Mouille, G.; Höfte, H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr. Biol. 2012, 22, 1732–1737. [Google Scholar] [CrossRef]
- Hoeberichts, F.; van Doorn, W.G.; Vorst, O.; Hall, R.D.; van Wordragen, M.F. Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J. Exp. Bot. 2007, 58, 2873–2885. [Google Scholar] [Green Version]
- Wang, H.; Stier, G.; Lin, J.; Liu, G.; Zhang, Z.; Chang, Y.; Reid, M.; Jiang, C. Transcriptome Changes Associated with Delayed Flower Senescence on Transgenic Petunia by Inducing Expression of etr1-1, a Mutant Ethylene Receptor. PLoS ONE 2013, 8, e65800. [Google Scholar] [CrossRef]
- Paniagua, C.; Posé, S.; Morris, V.J.; Kirby, A.R.; Quesada, M.A.; Mercado, J.A. Fruit softening and pectin disassembly: An overview of nanostructural pectin modifications assessed by atomic force microscopy. Ann. Bot. 2014, 114, 1375–1383. [Google Scholar] [CrossRef]
- Giovane, A.; Balestrieri, C.; Quagliuolo, L.; Castaldo, D.; Servillo, L. A glycoprotein inhibitor of pectin methylesterase in kiwi fruit. Purification by affinity chromatography and evidence of a ripening-related precursor. Eur. J. Biochem. 1995, 233, 926–929. [Google Scholar] [CrossRef]
- Irifune, K.; Nishida, T.; Egawa, H.; Nagatani, A. Pectin methylesterase inhibitor cDNA from kiwi fruit. Plant Cell Rep. 2004, 23, 333–338. [Google Scholar] [CrossRef]
- Damm, T.; Commandeur, U.; Fischer, R.; Usadel, B.; Klose, H. Improving the utilization of lignocellulosic biomass by polysaccharide modification. Process Biochem. 2016, 51, 288–296. [Google Scholar] [CrossRef]
- Francocci, F.; Bastianelli, E.; Lionetti, V.; Ferrari, S.; De Lorenzo, G.; Bellincampi, D.; Cervone, F. Analysis of pectin mutants and natural accessions of Arabidopsis highlights the impact of de-methyl-esterified homogalacturonan on tissue saccharification. Biotechnol. Biofuels 2013, 6, 4–11. [Google Scholar] [CrossRef]
- Castaldo, D.; Lovoi, A.; Quagliuolo, L.; Servillo, L.; Balestrieri, C.; Giovane, A. Orange juices and concentrates stabilization by a proteic inhibitor of pectin methylesterase. J. Food Sci. 1991, 56, 1632–1634. [Google Scholar] [CrossRef]
- Jolie, R.P.; Duvetter, T.; Vandevenne, E.; Van Buggenhout, S.; Van Loey, A.M.; Hendrickx, M.E. A pectin-methylesterase-inhibitor-based molecular probe for in Situ detection of plant pectin methylesterase activity. J. Agric. Food Chem. 2010, 58, 5449–5456. [Google Scholar] [CrossRef]
- Zocca, F.; Lomolino, G.; Curioni, A.; Spettoli, P.; Lante, A. Detection of pectinmethylesterase activity in presence of methanol during grape pomace storage. Food Chem. 2007, 102, 59–65. [Google Scholar] [CrossRef]
- Zocca, F.; Lomolino, G.; Spettoli, P.; Lante, A. A Study on the relationship between the volatile composition of moscato and prosecco grappa and enzymatic activities involved in its production. J. Inst. Brew. 2008, 114, 262–269. [Google Scholar] [CrossRef]
- Lante, A.; Zocca, F.; Spettoli, P.; Lomolino, G.; Raiola, A.; Bellincampi, D.; Lionetti, V.; Giovane, A.; Camardella, L. Use of a Protein Inhibitor of Pectin Methylesterase for Reducing Methanol Formation in Grape Must and Marc, and Process Therefor. It. Patent WO2008104555A1, 4 September 2008. [Google Scholar]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19102878
Wormit A, Usadel B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). International Journal of Molecular Sciences. 2018; 19(10):2878. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19102878
Chicago/Turabian StyleWormit, Alexandra, and Björn Usadel. 2018. "The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs)" International Journal of Molecular Sciences 19, no. 10: 2878. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19102878