Reaction with Proteins of a Five-Coordinate Platinum(II) Compound
Abstract
:1. Introduction
2. Results and Discussion
2.1. Spectroscopic Studies
2.2. Crystallographic Studies
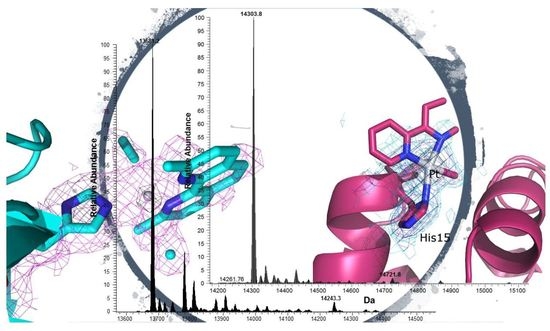
2.2.1. Structure of the HEWL Adduct
2.2.2. Structure of the RNase A Adduct
2.3. Mass Spectrometry Studies
3. Materials and Methods
3.1. Materials
3.2. Spectrophotometric Data
3.3. Crystallization, X-ray Diffraction Data Collection
3.4. Structure Solution and Refinement
3.5. Electrospray Ionization Mass Spectrometry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5C | pentacoordinate |
| a.u. | asymmetric unit |
| CA | carbon alpha |
| cisplatin | cis-Pt(NH3)2Cl2 |
| dmphen | 2,9-dimethyl-1,10-phenanthroline |
| DMSO | dimethylsulfoxide |
| DNA | deoxyribonucleic acid |
| e.d. | electron density |
| ESI MS | electrospray ionization mass spectrometry |
| HEWL | hen egg white lysozyme |
| Im | imidazole |
| Me | methyl |
| MLCT | metal–ligand charge transfer |
| NMR | nuclear magnetic resonance |
| olefin | dimethylfumarate |
| PDB | Protein Data Bank |
| PEG | polyethylene glycol |
| tbp geometry | trigonal bi-pyramidal geometry |
| rmsd | root mean square deviation |
| RNase A | bovine pancreatic ribonuclease |
| UV–Vis | ultraviolet–visible |
References
- Wheate, N.J.; Walker, S.; Craig, G.E.; Oun, R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010, 39, 8113–8127. [Google Scholar] [CrossRef] [PubMed]
- Galanski, M. Recent developments in the field of anticancer platinum complexes. Recent Pat Anti-Cancer Drug Discov. 2006, 1, 285–295. [Google Scholar] [CrossRef]
- Galanski, M.; Jakupec, M.A.; Keppler, B.K. Update of the preclinical situation of anticancer platinum complexes: Novel design strategies and innovative analytical approaches. Curr. Med. Chem. 2005, 12, 2075–2094. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, E.R.; Lippard, S.J. Structure, Recognition, and Processing of Cisplatin-DNA Adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.M.; Lippard, S.J. Cisplatin: From DNA damage to cancer chemotherapy. Prog. Nucleic Acid Res. Mol. Biol. 2001, 67, 93–130. [Google Scholar] [PubMed]
- Arnesano, F.; Natile, G. Mechanistic insight into the cellular uptake and processing of cisplatin 30 years after its approval by FDA. Coord. Chem. Rev. 2009, 253, 2070–2081. [Google Scholar] [CrossRef]
- Miller, R.P.; Tadagavadi, R.K.; Ramesh, G.; Reeves, W.B. Mechanisms of Cisplatin nephrotoxicity. Toxins 2010, 2, 2490–2518. [Google Scholar] [CrossRef]
- Messori, L.; Merlino, A. Cisplatin binding to proteins: A structural perspective. Coord. Chem. Rev. 2016, 315, 67–89. [Google Scholar] [CrossRef] [Green Version]
- Che, C.M.; Siu, F.M. Metal complexes in medicine with a focus on enzyme inhibition. Curr. Opin. Chem. Biol. 2010, 14, 255–261. [Google Scholar] [CrossRef]
- Casini, A.; Reedijk, J. Interactions of anticancer Pt compounds with proteins: An overlooked topic in medicinal inorganic chemistry? Chem. Sci. 2012, 3, 3135–3144. [Google Scholar] [CrossRef]
- Pinato, O.; Musetti, C.; Sissi, C. Pt-based drugs: The spotlight will be on proteins. Metallomics 2014, 6, 380–395. [Google Scholar] [CrossRef] [PubMed]
- Arnesano, F.; Natile, G. “Platinum on the road”: Interactions of antitumoral cisplatin with proteins. Pure Appl. Chem. 2008, 80, 2715–2725. [Google Scholar] [CrossRef]
- Cubo, L.; Hambley, T.W.; Sanz Miguel, P.J.; Carnero, A.; Navarro-Ranninger, C.; Quiroga, A.G. The preparation and characterization of trans-platinum(IV) complexes with unusually high cytotoxicity. Dalton Trans. 2011, 40, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.D.; Mellor, H.R.; Callaghan, R.; Hambley, T.W. Basis for design and development of platinum(IV) anticancer complexes. J. Med. Chem. 2007, 50, 3403–3411. [Google Scholar] [CrossRef] [PubMed]
- Reithofer, M.R.; Valiahdi, S.M.; Jakupec, M.A.; Arion, V.B.; Egger, A.; Galanski, M.; Keppler, B.K. Novel di- and tetracarboxylatoplatinum(IV) complexes. Synthesis, characterization, cytotoxic activity, and DNA platination. J. Med. Chem. 2007, 50, 6692–6699. [Google Scholar] [PubMed]
- Billecke, C.; Finniss, S.; Tahash, L.; Miller, C.; Mikkelsen, T.; Farrell, N.P.; Bögler, O. Polynuclear platinum anticancer drugs are more potent than cisplatin and induce cell cycle arrest in glioma. Neuro-Oncology 2006, 8, 215–226. [Google Scholar] [CrossRef]
- Bednarski, P.J.; Mackay, F.S.; Sadler, P.J. Photoactivatable platinum complexes. Anticancer Agents Med. Chem. 2007, 7, 75–93. [Google Scholar] [CrossRef]
- Cucciolito, M.E.; D’Amora, A.; De Feo, G.; Ferraro, G.; Giorgio, A.; Petruk, G.; Monti, D.M.; Merlino, A.; Ruffo, F. Five-Coordinate Platinum(II) Compounds Containing Sugar Ligands: Synthesis, Characterization, Cytotoxic Activity, and Interaction with Biological Macromolecules. Inorg. Chem. 2018, 57, 3133–3143. [Google Scholar] [CrossRef]
- Cucciolito, M.E.; De Luca Bossa, F.; Esposito, R.; Ferraro, G.; Iadonisi, A.; Petruk, G.; D’Elia, L.; Romanetti, C.; Traboni, S.; Tuzi, A.; et al. C-Glycosylation in platinum-based agents: A viable strategy to improve cytotoxicity and selectivity. Inorg. Chem. Front. 2018, 5, 2921–2933. [Google Scholar] [CrossRef]
- Albano, V.G.; Natile, G.; Panunzi, A. Five-coordinate alkene complexes of palladium(II) and platinum(II). Coord. Chem. Rev. 1994, 133, 67–114. [Google Scholar] [CrossRef]
- Maresca, L.; Natile, G. Five-Coordination in Platinum(II) and Palladium(II) Chemistry. Comment Inorg. Chem. 1993, 14, 349–366. [Google Scholar] [CrossRef]
- Benedetti, M.; Lamacchia, V.; Antonucci, D.; Papadia, P.; Pacifico, C.; Natile, G.; Fanizzi, F.P. Insertion of alkynes into Pt-X bonds of square planar [PtX2(N^N)] (X = Cl, Br, I) complexes. Dalton Trans. 2014, 43, 8826–8834. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.; Neugebauer, M.; Krest, A.; Lüning, A.; Garbe, S.; Arefyeva, N.; Schlörer, N. Five Coordinate Platinum(II) in [Pt(bpy)(cod)(Me)][SbF6]: A Structural and Spectroscopic Study. Inorganics 2015, 3, 118–138. [Google Scholar] [CrossRef]
- Carpinelli, P.; Bartolucci, S.; Ruffo, F. Synthesis and biological activity of five-coordinate platinum(II) complexes including organotin fragments. Anticancer Drug Des. 1995, 10, 43–49. [Google Scholar] [PubMed]
- Frezza, M.; Dou, Q.P.; Xiao, Y.; Samouei, H.; Rashidi, M.; Samari, F.; Hemmateenejad, B. In vitro and in vivo antitumor activities and DNA binding mode of five coordinated cyclometalated organoplatinum(II) complexes containing biphosphine ligands. J. Med. Chem. 2011, 54, 6166–6176. [Google Scholar] [CrossRef] [PubMed]
- Merlino, A. Interactions between proteins and Ru compounds of medicinal interest: A structural perspective. Coord. Chem. Rev. 2016, 326, 111–134. [Google Scholar] [CrossRef]
- Messori, L.; Marzo, T.; Gabbiani, C.; Valdes, A.A.; Quiroga, A.G.; Merlino, A. Peculiar features in the crystal structure of the adduct formed between cis-PtI2(NH3)2 and hen egg white lysozyme. Inorg. Chem. 2013, 52, 13827–13829. [Google Scholar] [CrossRef]
- Helliwell, J.R.; Tanley, S.W. The crystal structure analysis of the relative binding of cisplatin and carboplatin in a mixture with histidine in a protein studied at 100 and 300 K with repeated X-ray irradiation. Acta Crystallogr. D 2013, 69, 121–125. [Google Scholar] [CrossRef]
- Messori, L.; Merlino, A. Ruthenium metalation of proteins: The X-ray structure of the complex formed between NAMI-A and hen egg white lysozyme. Dalton Trans. 2014, 43, 6128–6131. [Google Scholar] [CrossRef]
- Messori, L.; Marzo, T.; Merlino, A. The X-ray structure of the complex formed in the reaction between oxaliplatin and lysozyme. Chem. Commun. 2014, 50, 8360–8362. [Google Scholar] [CrossRef]
- Vergara, A.; Russo Krauss, I.; Montesarchio, D.; Paduano, L.; Merlino, A. Investigating the ruthenium metalation of proteins: X-ray structure and Raman microspectroscopy of the complex between RNase A and AziRu. Inorg. Chem. 2013, 52, 10714–10716. [Google Scholar] [CrossRef] [PubMed]
- Gidney, P.M.; Gillard, R.D.; Heaton, B.T. Solvent effects on the electronic spectra of some 2,2‘-Bipyridyl Palladiurn(II) and Platinum(II) Complexes. J. Chem. Soc. Dalton Trans. 1973, 132–134. [Google Scholar] [CrossRef]
- Miskowski, V.M.; Houlding, V.H. Electronic spectra and photophysics of Platinum(II) complexes with a-diimine ligands. Solid-state effects. 1. Monomers and Ligand Dimers. Inorg. Chem. 1989, 28, 1529–1533. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Messori, L.; Cinellu, M.A.; Marasco, D.; Sirignano, R.; Merlino, A. Interactions of gold-based drugs with proteins: The structure and stability of the adduct formed in the reaction between lysozyme and the cytotoxic gold(III) compound Auoxo3. Dalton Trans. 2014, 43, 17483–17488. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Cinellu, M.A.; Merlino, A.; Messori, L.; Cinellu, M.A.; Merlino, A. Protein gold-based drug recognition:3D structure of the complex formed when Lysozyme reacts with Aubipyc. ACS Med. Chem. Lett. 2014, 5, 1110–1113. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Ferraro, G.; Pica, A.; Marquez, J.A.; Helliwell, J.R.; Merlino, A. Principles and methods used to grow and optimize crystals of protein-metallodrug adducts, to determine metal binding sites and to assign metal ligands. Metallomics 2017, 9, 1534–1547. [Google Scholar] [CrossRef] [PubMed]
- Tanley, S.W.; Diederichs, K.; Kroon-Batenburg, L.M.; Levy, C.; Schreurs, A.M.; Helliwell, J.R. Carboplatin binding to histidine. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 1135–1142. [Google Scholar] [CrossRef]
- Tanley, S.W.; Starkey, L.V.; Lamplough, L.; Kaenket, S.; Helliwell, J.R. The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15. Acta Cryst. F Struct. Biol. Commun. 2014, 70, 1132–1134. [Google Scholar] [CrossRef]
- Marasco, D.; Messori, L.; Marzo, T.; Merlino, A. Oxaliplatin vs. cisplatin: Competition experiments on their binding to lysozyme. Dalton Trans. 2015, 4, 10392–10398. [Google Scholar] [CrossRef]
- Mügge, C.; Marzo, T.; Massai, L.; Hildebrandt, J.; Ferraro, G.; Rivera-Fuentes, P.; Metzler-Nolte, N.; Merlino, A.; Messori, L.; Weigand, W. Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding. Inorg. Chem. 2015, 54, 8560–8570. [Google Scholar] [CrossRef]
- Ferraro, G.; Pica, A.; Russo Krauss, I.; Pane, F.; Amoresano, A.; Merlino, A. Effect of temperature on the interaction of cisplatin with the model protein hen egg white lysozyme. J. Biol. Inorg. Chem. 2016, 21, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Picone, D.; Donnarumma, F.; Ferraro, G.; Gotte, G.; Fagagnini, A.; Butera, G.; Donadelli, M.; Merlino, A. A comparison study on RNase A oligomerization induced by cisplatin, carboplatin and oxaliplatin. J. Inorg. Biochem. 2017, 173, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Marzo, T.; Merlino, A. Interactions of carboplatin and oxaliplatin with proteins: Insights from X-ray structures and mass spectrometry studies of their ribonuclease A adducts. J. Inorg. Biochem. 2015, 153, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Vitagliano, L.; Merlino, A.; Zagari, A.; Mazzarella, L. Reversible substrate-induced domain motions in ribonuclease A. Proteins 2002, 46, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Vitagliano, L.; Merlino, A.; Zagari, A.; Mazzarella, L. Productive and non-productive binding to ribonuclease A: X-ray structure of two complexes with uridylyl(2’,5’)guanosine. Protein Sci. 2000, 9, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Otwinoski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Cryst. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Vaney, M.C.; Maignan, S.; Ries-Kautt, M.; Ducruix, A. High-resolution structure (1.33 Å) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission. Acta Crystallogr. Sect. 1996, 52, 505–517. [Google Scholar] [CrossRef]
- Messori, L.; Merlino, A. Protein metalation by metal-based drugs: X-ray crystallography and mass spectrometry studies. Chem. Commun. (Camb.) 2017, 53, 11622–11633. [Google Scholar] [CrossRef]
- Merlino, A.; Marzo, T.; Messori, L. Protein Metalation by Anticancer Metallodrugs: A Joint ESI MS and XRD Investigative Strategy. Chem. Eur. J. 2017, 23, 6942–6947. [Google Scholar] [CrossRef] [PubMed]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferraro, G.; Marzo, T.; Cucciolito, M.E.; Ruffo, F.; Messori, L.; Merlino, A. Reaction with Proteins of a Five-Coordinate Platinum(II) Compound. Int. J. Mol. Sci. 2019, 20, 520. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030520
Ferraro G, Marzo T, Cucciolito ME, Ruffo F, Messori L, Merlino A. Reaction with Proteins of a Five-Coordinate Platinum(II) Compound. International Journal of Molecular Sciences. 2019; 20(3):520. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030520
Chicago/Turabian StyleFerraro, Giarita, Tiziano Marzo, Maria Elena Cucciolito, Francesco Ruffo, Luigi Messori, and Antonello Merlino. 2019. "Reaction with Proteins of a Five-Coordinate Platinum(II) Compound" International Journal of Molecular Sciences 20, no. 3: 520. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030520