Multipoint TvDAAO Mutants for Cephalosporin C Bioconversion
Abstract
:1. Introduction
2. Results
2.1. Comparison of Two Methods for Determination of TvDAAO Concentration
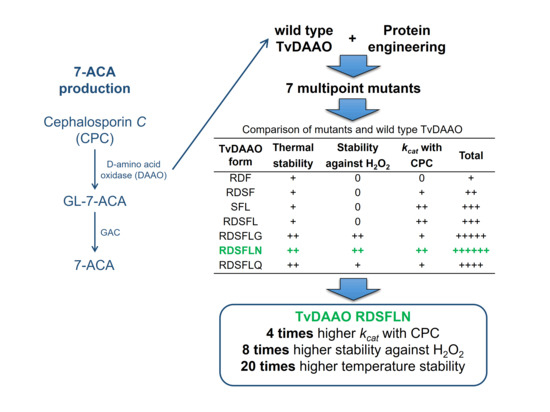
2.2. Preparation of Miltipoint TvDAAO Mutants
- E32R/F33D/C108F (abbreviation used in the manuscript RDF)
- E32R/F33D/F54S/C108F (RDSF)
- F54S/C108F/M156L (SFL)
- E32R/F33D/F54S/C108F/M156L (RDSFL)
- E32R/F33D/F54S/C108F/M156L/C298G (RDSFLG)
- E32R/F33D/F54S/C108F/M156L/C298N (RDSFLN)
- E32R/F33D/F54S/C108F/M156L/C298Q (RDSFLQ)
2.3. Thermal Stability of TvDAAO Mutants
2.4. Stability of Mutant TvDAAOs Against Hydrogen Peroxide
2.5. Kinetic Properties of Mutant TvDAAOs with d-Amino Acids
2.6. Kinetic Properties of Mutant TvDAAOs with Cephalosporin C
3. Discussion
4. Materials and Methods
4.1. Preparation of TvDAAO Mutants
4.2. Kinetic Assay
4.3. Thermal and Oxidation Stability
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DAAO | d-amino acid oxidase |
| 7-ACA | 7-aminocephalosporanic acid |
| CPC | Cephalosporin C |
| GAC | Glutaryl-7-ACA acylase |
| KA-7-ACA | α-keto adipil-7-ACA |
| GL-7-ACA | Glutaryl-7-ACA |
| TvDAAO | d-amino acid oxidase from yeast Trigonopsis variabilis |
| BSA | Bovine serum albumin |
| wt-TvDAAO | Wild-type TvDAAO |
| RDF | Mutant TvDAAO with amino acid changes E32R/F33D/C108F |
| RDSF | Mutant TvDAAO with amino acid changes E32R/F33D/F54S/C108F |
| SFL | Mutant TvDAAO with amino acid changes F54S/C108F/M156L |
| RDSFL | Mutant TvDAAO with amino acid changes E32R/F33D/F54S/C108F/M156L |
| RDSFLG | Mutant TvDAAO with amino acid changes E32R/F33D/F54S/C108F/M156L/C298G |
| RDSFLN | Mutant TvDAAO with amino acid changes E32R/F33D/F54S/C108F/M156L/C298N |
| RDSFLQ | Mutant TvDAAO with amino acid changes E32R/F33D/F54S/C108F/M156L/C298Q |
| SDS-PAGE | Polyacrylamide gel electrophoresis in presence of sodium dodecyl sulfate |
| KPB | Potassium phosphate buffer |
| ABTS | 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid |
| HPLC | High pressure liquid chromatography |
| OPD | o-phenylenediamine |
| HRP | Horseradish peroxidase |
Appendix A
| Amino Acid | wt-TvDAAO | TvDAAO RDF | TvDAAO RDSF | TvDAAO SFL | ||||
|---|---|---|---|---|---|---|---|---|
| KM, mM | kcat, s−1 | KM, mM | kcat, s−1 | KM, mM | kcat, s−1 | KM, mM | kcat, s−1 | |
| d-Met | 0.46 | 81 | 2.7 | 981 | 1.3 | 37 | 1.2 | 39 |
| d-Ala | 17 | 109 | 51 | 31 | 6.4 | 3.1 | - | - |
| d-Val | 14 | 85 | 39 | 37 | 5.1 | 2.5 | - | - |
| d-Leu | 0.40 | 29 | 0.58 | 21 | 0.09 | 12 | 0.1 | 17 |
| d-Phe | 0.37 | 27 | 1.1 | 63 | 1.7 | 92 | 1.3 | 79 |
| d-Tyr | 0.45 | 22 | 0.18 | 11 | 0.36 | 52 | 0.14 | 39 |
| d-Trp | 0.49 | 42 | 0.79 | 28 | 1.0 | 23 | 0.74 | 19 |
| d-Asn | 23 | 62 | 161 | 107 | 4.7 | 68 | 5.2 | 83 |
| d-Lys | 29 | 3.5 | 48 | 7.3 | 12 | 8 | - | - |
| d-Ser | 37 | 20 | 32 | 5.5 | - | - | - | - |
| d-Thr | 11 | 1.8 | - 2 | - | - | - | - | - |
| Amino Acid | TvDAAO RDSFL | TvDAAO RDSFLG | TvDAAO RDSFLN | TvDAAO RDSFLQ | ||||
|---|---|---|---|---|---|---|---|---|
| KM, mM | kcat, s−1 | KM, mM | kcat, s−1 | KM, mM | kcat, s−1 | KM, mM | kcat, s−1 | |
| d-Met | 1.1 | 33 | 1.1 | 28 | 1.4 | 41 | 0.98 | 33 |
| d-Ala | 6.11 | 2.3 | 4.9 | 4.2 | 3.7 | 4.9 | 1.8 | 4.3 |
| d-Val | 4.0 | 2.6 | 3.6 | 3.8 | 4.5 | 3.7 | 3.1 | 3.0 |
| d-Leu | 0.09 | 14 | 0.08 | 18 | 0.11 | 18 | 0.09 | 14 |
| d-Phe | 0.33 | 48 | 0.37 | 37 | 0.77 | 55 | 1.2 | 31 |
| d-Tyr | 0.19 | 44 | 0.17 | 28 | 0.21 | 42 | 0.17 | 14 |
| d-Trp | 0.61 | 18 | 0.81 | 14 | 0.87 | 22 | 0.58 | 6.7 |
| d-Asn | 4.3 | 69 | 2.9 | 58 | 3.4 | 68 | 3.7 | 61 |
| d-Lys | 15 | 8.6 | 8.9 | 7.6 | 17 | 12 | 14 | 6.6 |
| d-Ser | 9.0 | 0.98 | - 2 | - | 9.5 | 2.2 | - | - |
| d-Thr | 9.8 | 0.47 | - | - | - | - | - | - |
References
- Pollegioni, L.; Diederichs, K.; Molla, G.; Umhau, S.; Welte, W.; Ghisla, S.; Pilone, M.S. Yeast D-amino acid oxidase: Structural basis of its catalytic properties. J. Mol. Biol. 2002, 324, 535–546. [Google Scholar] [CrossRef]
- Tishkov, V.I.; Khoronenkova, S.V. D-Amino acid oxidase: Structure, catalytic mechanism, and practical application. Biochemistry 2005, 70, 40–54. [Google Scholar] [CrossRef]
- Khoronenkova, S.V.; Tishkov, V.I. D-amino acid oxidase: Physiological role and applications. Biochemistry 2008, 73, 1511–1518. [Google Scholar] [CrossRef]
- Pollegioni, L.; Molla, G.; Sacchi, S.; Rosini, E.; Verga, R.; Pilone, M.S. Properties and applications of microbial D-amino acid oxidases: Current state and perspectives. Appl. Microbiol. Biotechnol. 2008, 78, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.S.; Giesecke, U.; Reichert, A.; Minas, W. Industrial enzymatic production of cephalosporin-based beta-lactams. Adv. Biochem. Eng. Biotechnol. 2004, 88, 179–215. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T. 7-Aminocephalosporanic acid—Chemical versus enzymatic production process. In Asymmetric Catalysis on Industrial Scale: Challenges, Approaches and Solutions; Wiley-VCH: Weinheim, Germany, 2004; pp. 117–130. [Google Scholar] [CrossRef]
- Pilone, M.S.; Pollegioni, L. D-amino acid oxidase as an industrial biocatalyst. Biocatal. Biotransform. 2002, 20, 145–159. [Google Scholar] [CrossRef]
- Lopez-Gallego, F.; Batencor, L.; Hidalgo, A.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. One-Pot Conversion of cephalosporin C to 7-aminocephalosporanic acid in the absence of hydrogen peroxide. Adv. Synth. Catal. 2005, 347, 1804–1810. [Google Scholar] [CrossRef]
- Conti, G.; Pollegioni, L.; Rosini, E. One-pot conversion of cephalosporin C by using an optimized two-enzyme process. Catal. Sci. Technol. 2015, 5, 1854–1863. [Google Scholar] [CrossRef]
- Tan, Q.; Qiu, J.; Luo, X.; Zhang, Y.; Liu, Y.; Chen, Y.; Yuan, J.; Liao, W. Progress in one-pot bioconversion of cephalosporin C to 7-aminocephalosporanic acid. Curr. Pharm. Biotechnol. 2018, 19, 30–42. [Google Scholar] [CrossRef]
- Ishii, Y.; Saito, Y.; Fujimura, T.; Sasaki, H.; Noguchi, Y.; Yamada, H.; Niwa, M.; Shimomura, K. High-level production, chemical modification and site-directed mutagenesis of a cephalosporin C acylase from Pseudomonas strain N176. Eur. J. Biochem. 1995, 230, 773–778. [Google Scholar] [CrossRef]
- Saito, Y.; Fujimura, T.; Ishii, Y.; Noguchi, Y.; Miura, T.; Niwa, M.; Shimomura, K. Oxidative modification of a cephalosporin C acylase from Pseudomonas strain N176 and site-directed mutagenesis of the gene. Appl. Environ. Microbiol. 1996, 62, 2919–2925. [Google Scholar] [PubMed]
- Pollegioni, L.; Caldinelli, L.; Molla, G.; Sacchi, S.; Pilone, M.S. Catalytic properties of D-amino acid oxidase in cephalosporin C bioconversion: A comparison between proteins from different sources. Biotechnol. Prog. 2004, 20, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.V.; Golubev, I.V.; Khoronenkova, S.V.; Tishkov, V.I. Mutant D-amino acid oxidase with higher catalytic efficiency toward D-amino acids with bulky side chains. Russ. Chem. Bull. 2012, 61, 1489–1496. [Google Scholar] [CrossRef]
- Wong, K.S.; Fong, W.P.; Tsang, P.W. A single Phe54Tyr substitution improves the catalytic activity and thermostability of Trigonopsis variabilis D-amino acid oxidase. New Biotechnol. 2010, 27, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Schrader, T.; Andreesen, J.R. Evidence for the functional importance of Cys298 in D-amino acid oxidase from Trigonopsis variabilis. Eur. J. Biochem. 1993, 218, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Schrader, T.; Andreesen, J.R. Studies on the inactivation of the flavoprotein D-amino acid oxidase from Trigonopsis variabilis. Appl. Microbiol. Biotechnol. 1996, 45, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Slavica, A.; Dib, I.; Nidetzky, B. Single-site oxidation, cysteine 108 to cysteine sulfinic acid, in D-amino acid oxidase from Trigonopsis variabilis and its structural and functional consequences. Appl. Environ. Microbiol. 2005, 71, 8061–8068. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Kratzer, R.; Schiller, M.; Slavica, A.; Rechberger, G.; Kollroser, M.; Nidetzky, B. The role of Cys108 in Trigonopsis variabilis D-amino acid oxidase examined through chemical oxidation studies and point mutations C108S and C108D. Biochim. Biophys. Acta 2010, 1804, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Khoronenkova, S.V.; Tishkov, V.I. Recombinant D-amino acid oxidase with improved properties. Chin. J. Biotechnol. 2008, 24, 2122–2124. [Google Scholar] [CrossRef]
- Tishkov, V.I.; Khoronenkova, S.V.; Savina, L.I. Mutant D-amino Acid Oxidases. Russian Patent RU 2,362,806, 27 July 2009. [Google Scholar]
- Cherskova, N.V.; Khoronenkova, S.V.; Panteleev, M.A.; Tishkov, V.I. Site-directed mutagenesis of D-amino acid oxidase from yeast Trigonopsis variabilis. J. Biotechnol. 2010, 150, 442. [Google Scholar] [CrossRef]
- Ju, S.S.; Lin, L.L.; Chien, H.R.; Hsu, W.H. Substitution of the critical methionine residues in Trigonopsis variabilis D-amino acid oxidase with leucine enhances its resistance to hydrogen peroxide. FEMS Microbiol. Lett. 2000, 186, 215–219. [Google Scholar] [CrossRef]
- Atroshenko, D.L.; Golubev, I.V.; Savin, S.S.; Tishkov, V.I. Influence of Met/Leu amino acid changes on catalytic properties and oxidative and thermal stability of yeast D-amino acid oxidase. Mosc. Univ. Chem. Bull. 2016, 71, 243–252. [Google Scholar] [CrossRef]
- Tishkov, V.I.; Pometun, A.A.; Stepashkina, A.V.; Fedorchuk, V.V.; Zarubina, S.A.; Kargov, I.S.; Atroshenko, D.L.; Parshin, P.D.; Shelomov, M.D.; Kovalevski, R.P.; et al. Rational design of practically important enzymes. Mosc. Univ. Chem. Bull. 2018, 73, 1–6. [Google Scholar] [CrossRef]
- Golubev, I.V. Structural and functional studies of yeast D-amino acid oxidase with rational design approach. Ph.D. Dissertation, Lomonosov Moscow State University, Moscow, Russia, 16 December 2014. [Google Scholar]
- Golubev, I.V.; Komarova, N.V.; Ryzhenkova, K.V.; Chubar, T.A.; Savin, S.S.; Tishkov, V.I. Study of the structure-function-stability relationships in yeast D-amino acid oxidase: Hydrophobization of alpha-helices. Acta Nat. 2014, 6, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Dib, I.; Slavica, A.; Riethorst, W.; Nidetzky, B. Thermal inactivation of D-amino acid oxidase from Trigonopsis variabilis occurs via three parallel paths of irreversible denaturation. Biotechnol. Bioeng. 2006, 94, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Schrader, T.; Andreesen, J.R. Properties and chemical modification of D-amino acid oxidase from Trigonopsis variabilis. Arch. Microbiol. 1996, 165, 41–47. [Google Scholar] [CrossRef]
- Wang, S.J.; Yu, C.Y.; Kuan, I.C. Stabilization of native and double D-amino acid oxidases from Rhodosporidium toruloides and Trigonopsis variabilis by immobilization on streptavidin-coated magnetic beads. Biotechnol. Lett. 2008, 30, 1973–1981. [Google Scholar] [CrossRef]
- Savin, S.S.; Chernyshov, I.V.; Tishkov, V.I.; Khoronenkova, S.V. Substrate specifity of D-amino acid oxidase from yeast Trigonopsis variabilis expressed in E. coli cells. Mosc. Univ. Chem. Bull. 2006, 61, 13–19. [Google Scholar]





| TvDAAO Form | Yield, % | Specific Activity 1, (U/mg) |
|---|---|---|
| wild-type | 44 | 140 |
| RDF | 44 | 149 |
| RDSF | 43 | 47 |
| SFL | 61 | 63 |
| RDSFL | 95 | 50 |
| RDSFLG | 90 | 55 |
| RDSFLN | 72 | 66 |
| RDSFLQ | 82 | 46 |
| TvDAAO form | Temperature (°C) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 50 | 52 | 54 | 56 | 58 | 60 | 62 | 64 | 66 | |
| wild-type | 73 | 47 | 23 | 7.2 | 4.7 | 2.7 | - | - | - |
| RDF | - | - | 150 | 42 | 19 | 8.9 | 3.1 | - | - |
| RDSF | - | - | 79 | 33 | 9.5 | 5.6 | 2.8 | - | - |
| SFL | - | - | - | 110 | 57 | 17 | 9.8 | - | - |
| RDSFL | - | - | - | - | 43 | 15 | 4.6 | 1.4 | - |
| RDSFLG | - | - | - | - | - | 41 | 27 | 6.5 | 2.6 |
| RDSFLN | - | - | - | - | - | 54 | 19 | 6.0 | 4.2 |
| RDSFLQ | - | - | - | - | - | 77 | 16 | 4.0 | - |
| TvDAAO Form | [H2O2], (M) | |
|---|---|---|
| 0.01 M | 0.1 M | |
| wild-type | 14 | 2.6 |
| RDF | - | 1.6 |
| RDSF | - | 1.8 |
| SFL | 17 | 1.7 |
| RDSFL | 9 | 1.5 |
| RDSFLG | 120 | 25 |
| RDSFLN | 120 | 22 |
| RDSFLQ | 90 | 18 |
| TvDAAO Form | KM, (mM) | kcat, (s−1) | kcat/KM, (s−1·mM−1) |
|---|---|---|---|
| wild-type | 1.4 ± 0.4 | 26 ± 3 | 18 ± 5 |
| RDF | 1.15 ± 0.081 | 25.5 ± 1.4 | 22 ± 2 |
| RDSF | 1.9 ± 0.4 | 88 ± 10 | 45 ± 11 |
| SFL | 1.9 ± 0.6 | 119 ± 7 | 60 ± 20 |
| RDSFL | 1.6 ± 0.3 | 106 ± 9 | 66 ± 12 |
| RDSFLG | 0.8 ± 0.2 | 65 ± 5 | 79 ± 15 |
| RDSFLN | 2.9 ± 0.5 | 121 ± 15 | 42 ± 8 |
| RDSFLQ | 1.4 ± 0.3 | 68 ± 0.5 | 50 ± 9 |
| Source | wt-TvDAAO Concentration | Reaction Condition | Measurement Methods | Catalytic Properties from Source | Recalculated Catalytic Properties 1 |
|---|---|---|---|---|---|
| This work | FAD extinction | pH 8.0, 30 °C 100 mM NaPB | Cephalosporin C consumption | KM = 1.4 mM kcat = 26 s−1 | KM = 1.4 mM kcat = 26 s−1 |
| [29] | Bradford | pH 8.0, 37 °C 0.21 mM O2 | Oxygen consumption | KM = 0.83 mM Specific activity = 46 U/mg (kcat = 30.2 s−1) | KM = 0.83 mM kcat = 49 s−1 |
| [15] | Bradford | pH 7.5, 22 °C 50 mM NaPB | GL-7-ACA production | KM = 1.6 mM kcat = 370 min−1 (kcat = 6.2 s−1) | KM = 1.6 mM kcat = 10 s−1 |
| [13] | FAD extinction | pH 8.5, 25 °C 0.2 mM FAD air saturated | Oxygen consumption | KM = 2.4 mM kcat = 4300 min−1 (kcat = 72 s−1) | KM = 2.4 mM kcat = 72 s−1 |
| [30] | Bradford | pH 8.0, 25 °C 100 mM KPB | OPD/HRP | KM = 9 mM kcat = 55 s−1 | KM = 9 mM kcat = 89 s−1 |
| TvDAAO Form | Thermal Stability | Oxidative Stability | kcat with CPC | Total |
|---|---|---|---|---|
| RDF | + | 0 | 0 | + |
| RDSF | + | 0 | + | ++ |
| SFL | + | 0 | ++ | +++ |
| RDSFL | + | 0 | ++ | +++ |
| RDSFLG | ++ | ++ | + | +++++ |
| RDSFLN | ++ | ++ | ++ | ++++++ |
| RDSFLQ | ++ | + | + | ++++ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atroshenko, D.L.; Shelomov, M.D.; Zarubina, S.A.; Negru, N.Y.; Golubev, I.V.; Savin, S.S.; Tishkov, V.I. Multipoint TvDAAO Mutants for Cephalosporin C Bioconversion. Int. J. Mol. Sci. 2019, 20, 4412. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184412
Atroshenko DL, Shelomov MD, Zarubina SA, Negru NY, Golubev IV, Savin SS, Tishkov VI. Multipoint TvDAAO Mutants for Cephalosporin C Bioconversion. International Journal of Molecular Sciences. 2019; 20(18):4412. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184412
Chicago/Turabian StyleAtroshenko, Denis L., Mikhail D. Shelomov, Sophia A. Zarubina, Nikita Y. Negru, Igor V. Golubev, Svyatoslav S. Savin, and Vladimir I. Tishkov. 2019. "Multipoint TvDAAO Mutants for Cephalosporin C Bioconversion" International Journal of Molecular Sciences 20, no. 18: 4412. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184412