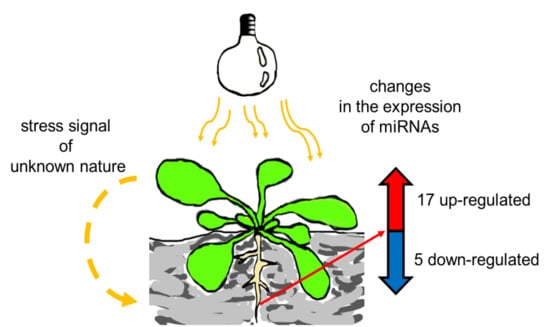
Exposure to High-Intensity Light Systemically Induces Micro-Transcriptomic Changes in Arabidopsis thaliana Roots
Abstract
:1. Introduction
2. Results
2.1. HL-Induced Transcriptional Changes in A. thaliana Roots
2.2. HL-Triggered Systemic Changes in the Expression of miRNAs
2.3. Stress Signal is Induced in Rosettes
2.4. Prediction of Potential Targets
3. Discussion
4. Material and Methods
4.1. Plant Material and Growing Conditions
4.2. High-Light Treatment
4.3. Root Separation
4.4. RNA Isolation
4.5. Preparation of Libraries and Micro-Transcriptomic Analysis
4.6. Expression of Marker Genes Determined by RT-qPCR
4.6.1. cDNA Synthesis
4.6.2. Quantitative PCR
4.7. Detection of Mature miRNAs Using Two-Tailed qPCR
4.7.1. cDNA Synthesis
4.7.2. Quantitative PCR
4.8. Detection of Mature miRNA Using Mir-X miRNA
4.8.1. cDNA Synthesis
4.8.2. Quantitative PCR
4.9. Target Transcript Prediction
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Van Gelderen, K.; Kang, C.; Pierik, R. Light Signaling, Root Development, and Plasticity. Plant Physiol. 2018, 176, 1049–1060. [Google Scholar] [CrossRef]
- Ruffel, S.; Gojon, A. Systemic nutrient signalling: On the road for nitrate. Nat. Plants 2017, 3, 17040. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef]
- Foyer, C.H.; Shigeoka, S. Understanding Oxidative Stress and Antioxidant Functions to Enhance Photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef]
- Fryer, M.J. The antioxidant effects of thylakoid Vitamin E (α-tocopherol). Plant Cell Environ. 1992, 15, 381–392. [Google Scholar] [CrossRef]
- Murchie, E.H.; Niyogi, K.K. Manipulation of Photoprotection to Improve Plant Photosynthesis. Plant Physiol. 2011, 155, 86–92. [Google Scholar] [CrossRef]
- Ware, M.A.; Belgio, E.; Ruban, A.V. Comparison of the protective effectiveness of NPQ in Arabidopsis plants deficient in PsbS protein and zeaxanthin. J. Exp. Bot. 2015, 66, 1259–1270. [Google Scholar] [CrossRef]
- Mo, M.; Yokawa, K.; Wan, Y.; Baluška, F. How and why do root apices sense light under the soil surface? Front. Plant Sci. 2015, 6, 775. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Yanagisawa, S. Light signalling-induced regulation of nutrient acquisition and utilisation in plants. Semin. Cell Dev. Biol. 2018, 83, 123–132. [Google Scholar] [CrossRef]
- Lee, H.-J.; Ha, J.-H.; Kim, S.-G.; Choi, H.-K.; Kim, Z.H.; Han, Y.-J.; Kim, J.-I.; Oh, Y.; Fragoso, V.; Shin, K.; et al. Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci. Signal. 2016, 9, ra106. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Ha, J.-H.; Park, C.-M. Underground roots monitor aboveground environment by sensing stem-piped light. Commun. Integr. Biol. 2016, 9, e1261769. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yao, Q.; Gao, X.; Jiang, C.; Harberd, N.P.; Fu, X. Shoot-to-Root Mobile Transcription Factor HY5 Coordinates Plant Carbon and Nitrogen Acquisition. Curr. Biol. 2016, 26, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S. Nutrient-related Long-Distance Signals: Common players and possible crosstalk. Plant Cell Physiol. 2018. [Google Scholar] [CrossRef]
- Aung, K.; Lin, S.-I.; Wu, C.-C.; Huang, Y.-T.; Su, C.-L.; Chiou, T.-J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.-R. PHO2, MicroRNA399, and PHR1 Define a Phosphate-Signaling Pathway in Plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [Green Version]
- Chiou, T.-J.; Aung, K.; Lin, S.-I.; Wu, C.-C.; Chiang, S.-F.; Su, C.-L. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef]
- Fujii, H.; Chiou, T.-J.; Lin, S.-I.; Aung, K.; Zhu, J.-K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005, 15, 2038–2043. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef]
- Kawashima, C.G.; Yoshimoto, N.; Maruyama-Nakashita, A.; Tsuchiya, Y.N.; Saito, K.; Takahashi, H.; Dalmay, T. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. Cell Mol. Biol. 2009, 57, 313–321. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Yu, D. Identification of Nitrogen Starvation-Responsive MicroRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef]
- Barciszewska-Pacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A.; et al. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015, 6, 410. [Google Scholar] [CrossRef]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef]
- Brant, E.J.; Budak, H. Plant Small Non-coding RNAs and Their Roles in Biotic Stresses. Front. Plant Sci. 2018, 9, 1038. [Google Scholar] [CrossRef] [Green Version]
- Islam, W.; Qasim, M.; Noman, A.; Adnan, M.; Tayyab, M.; Farooq, T.H.; Wei, H.; Wang, L. Plant microRNAs: Front line players against invading pathogens. Microb. Pathog. 2018, 118, 9–17. [Google Scholar] [CrossRef]
- Chien, P.-S.; Chiang, C.-B.; Wang, Z.; Chiou, T.-J. MicroRNA-mediated signaling and regulation of nutrient transport and utilization. Curr. Opin. Plant Biol. 2017, 39, 73–79. [Google Scholar] [CrossRef]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef]
- Yang, B.; Tang, J.; Yu, Z.; Khare, T.; Srivastav, A.; Datir, S.; Kumar, V. Light Stress Responses and Prospects for Engineering Light Stress Tolerance in Crop Plants. J. Plant Growth Regul. 2019. [Google Scholar] [CrossRef]
- Chung, P.J.; Park, B.S.; Wang, H.; Liu, J.; Jang, I.-C.; Chua, N.-H. Light-Inducible MiR163 Targets PXMT1 Transcripts to Promote Seed Germination and Primary Root Elongation in Arabidopsis. Plant Physiol. 2016, 170, 1772–1782. [Google Scholar] [CrossRef]
- Petrillo, E.; Herz, M.A.G.; Fuchs, A.; Reifer, D.; Fuller, J.; Yanovsky, M.J.; Simpson, C.; Brown, J.W.S.; Barta, A.; Kalyna, M.; et al. A chloroplast retrograde signal regulates nuclear alternative splicing. Science 2014, 344, 427–430. [Google Scholar] [CrossRef]
- Ruckle, M.E.; Burgoon, L.D.; Lawrence, L.A.; Sinkler, C.A.; Larkin, R.M. Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol. 2012, 159, 366–390. [Google Scholar] [CrossRef]
- Karpinski, S.; Reynolds, H.; Karpinska, B.; Wingsle, G.; Creissen, G.; Mullineaux, P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 1999, 284, 654–657. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Kruk, J.; Górecka, M.; Karpińska, B.; Karpiński, S. Evidence for Light Wavelength-Specific Photoelectrophysiological Signaling and Memory of Excess Light Episodes in Arabidopsis. Plant Cell 2010, 22, 2201–2218. [Google Scholar] [CrossRef]
- Asada, K. Production and Scavenging of Reactive Oxygen Species in Chloroplasts and Their Functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP Chloroplast Retrograde Pathway That Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef]
- Gordon, M.J.; Carmody, M.E.; Albrecht, V.; Pogson, B. Systemic and Local Responses to Repeated HL Stress-Induced Retrograde Signaling in Arabidopsis. Front. Plant Sci. 2013, 3, 303. [Google Scholar] [CrossRef] [Green Version]
- Karpinski, S.; Escobar, C.; Karpinska, B.; Creissen, G.; Mullineaux, P.M. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 1997, 9, 627–640. [Google Scholar]
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540. [Google Scholar] [CrossRef] [Green Version]
- Shao, N.; Duan, G.Y.; Bock, R. A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells. Plant Cell 2013, 25, 4209–4226. [Google Scholar] [CrossRef] [PubMed]
- Shumbe, L.; D’Alessandro, S.; Shao, N.; Chevalier, A.; Ksas, B.; Bock, R.; Havaux, M. METHYLENE BLUE SENSITIVITY 1 (MBS1) is required for acclimation of Arabidopsis to singlet oxygen and acts downstream of β-cyclocitral. Plant Cell Environ. 2017, 40, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Przybyla, D.; Op den Camp, R.; Kim, C.; Landgraf, F.; Lee, K.P.; Würsch, M.; Laloi, C.; Nater, M.; Hideg, E.; et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 2004, 306, 1183–1185. [Google Scholar] [CrossRef] [PubMed]
- Mühlenbock, P.; Szechyńska-Hebda, M.; Płaszczyca, M.; Baudo, M.; Mateo, A.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpiński, S. Chloroplast Signaling and LESION SIMULATING DISEASE1 Regulate Crosstalk between Light Acclimation and Immunity in Arabidopsis. Plant Cell 2008, 20, 2339–2356. [Google Scholar] [CrossRef]
- Mullineaux, P.; Ball, L.; Escobar, C.; Karpinska, B.; Creissen, G.; Karpinski, S. Are diverse signalling pathways integrated in the regulation of arabidopsis antioxidant defence gene expression in response to excess excitation energy? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1531–1540. [Google Scholar] [CrossRef]
- Rossel, J.B.; Wilson, P.B.; Hussain, D.; Woo, N.S.; Gordon, M.J.; Mewett, O.P.; Howell, K.A.; Whelan, J.; Kazan, K.; Pogson, B.J. Systemic and Intracellular Responses to Photooxidative Stress in Arabidopsis. Plant Cell 2007, 19, 4091–4110. [Google Scholar] [CrossRef]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubista, M. Two-tailed RT-qPCR: A novel method for highly accurate miRNA quantification. Nucleic Acids Res. 2017, 45, e144. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Finet, C.; Fourquin, C.; Vinauger, M.; Berne-Dedieu, A.; Chambrier, P.; Paindavoine, S.; Scutt, C.P. Parallel structural evolution of auxin response factors in the angiosperms. Plant J. 2010, 63, 952–959. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Williams, M.E.; Torabinejad, J.; Cohick, E.; Parker, K.; Drake, E.J.; Thompson, J.E.; Hortter, M.; Dewald, D.B. Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol. 2005, 138, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Meurer, J.; Lezhneva, L.; Amann, K.; Gödel, M.; Bezhani, S.; Sherameti, I.; Oelmüller, R. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell 2002, 14, 3255–3269. [Google Scholar] [CrossRef] [PubMed]
- Stokes, K.D.; McAndrew, R.S.; Figueroa, R.; Vitha, S.; Osteryoung, K.W. Chloroplast Division and Morphology Are Differentially Affected by Overexpression of FtsZ1 and FtsZ2 Genes in Arabidopsis. Plant Physiol. 2000, 124, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Barczak-Brzyżek, A.K.; Kiełkiewicz, M.; Gawroński, P.; Kot, K.; Filipecki, M.; Karpińska, B. Cross-talk between high light stress and plant defence to the two-spotted spider mite in Arabidopsis thaliana. Exp. Appl. Acarol. 2017, 73, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waszczak, C.; Kerchev, P.I.; Mühlenbock, P.; Hoeberichts, F.A.; Van Der Kelen, K.; Mhamdi, A.; Willems, P.; Denecker, J.; Kumpf, R.P.; Noctor, G.; et al. SHORT-ROOT Deficiency Alleviates the Cell Death Phenotype of the Arabidopsis catalase2 Mutant under Photorespiration-Promoting Conditions. Plant Cell 2016, 28, 1844–1859. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M.; Johnson, J.M.; Hieno, A.; Tokizawa, M.; Nomoto, M.; Tada, Y.; Godfrey, R.; Obokata, J.; Sherameti, I.; Yamamoto, Y.Y.; et al. High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 Levels Result in Accumulation of Reactive Oxygen Species in Arabidopsis thaliana Shoots and Roots. Mol. Plant 2015, 8, 1253–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutin, C.; Nussaume, L.; Moise, N.; Moya, I.; Kloppstech, K.; Havaux, M. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc. Natl. Acad. Sci. USA 2003, 100, 4921–4926. [Google Scholar] [CrossRef]
- Auge, G.A.; Perelman, S.; Crocco, C.D.; Sánchez, R.A.; Botto, J.F. Gene expression analysis of light-modulated germination in tomato seeds. New Phytol. 2009, 183, 301–314. [Google Scholar] [CrossRef]
- Rizza, A.; Boccaccini, A.; Lopez-Vidriero, I.; Costantino, P.; Vittorioso, P. Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytol. 2011, 190, 896–905. [Google Scholar] [CrossRef]
- Khan, A.; Goswami, K.; Sopory, S.K.; Sanan-Mishra, N. “Mirador” on the potential role of miRNAs in synergy of light and heat networks. Indian J. Plant Physiol. 2017, 22, 587–607. [Google Scholar] [CrossRef]
- Hernando, C.E.; Garcia, C.; Mateos, J.L. Casting Away the Shadows: Elucidating the Role of Light-mediated Posttranscriptional Control in Plants. Photochem. Photobiol. 2017, 93, 656–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.-P.; Su, Y.-S.; Chen, H.-C.; Chen, Y.-R.; Wu, C.-C.; Lin, W.-D.; Tu, S.-L. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 2014, 15, R10. [Google Scholar] [CrossRef] [PubMed]
- Achkar, N.P.; Cho, S.K.; Poulsen, C.; Arce, A.L.; Re, D.A.; Giudicatti, A.J.; Karayekov, E.; Ryu, M.Y.; Choi, S.W.; Harholt, J.; et al. A Quick HYL1-Dependent Reactivation of MicroRNA Production Is Required for a Proper Developmental Response after Extended Periods of Light Deprivation. Dev. Cell 2018, 46, 236–247.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Wang, J.-W.; Wang, L.-J.; Mao, Y.-B.; Cai, W.-J.; Xue, H.-W.; Chen, X.-Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Floková, K.; Pacurar, D.I.; Novák, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef]
- Schuman, M.C.; Meldau, S.; Gaquerel, E.; Diezel, C.; McGale, E.; Greenfield, S.; Baldwin, I.T. The Active Jasmonate JA-Ile Regulates a Specific Subset of Plant Jasmonate-Mediated Resistance to Herbivores in Nature. Front. Plant Sci. 2018, 9, 787. [Google Scholar] [CrossRef] [Green Version]
- Gläßer, C.; Haberer, G.; Finkemeier, I.; Pfannschmidt, T.; Kleine, T.; Leister, D.; Dietz, K.-J.; Häusler, R.E.; Grimm, B.; Mayer, K.F.X. Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Mol. Plant 2014, 7, 1167–1190. [Google Scholar] [CrossRef]
- Barczak-Brzyżek, A.; Kiełkiewicz, M.; Górecka, M.; Kot, K.; Karpińska, B.; Filipecki, M. Abscisic Acid Insensitive 4 transcription factor is an important player in the response of Arabidopsis thaliana to two-spotted spider mite (Tetranychus urticae) feeding. Exp. Appl. Acarol. 2017, 73, 317–326. [Google Scholar] [CrossRef]
- Itoh, R.D.; Fujiwara, M.T. Regulation of leucoplast morphology in roots. Plant Signal. Behav. 2010, 5, 856–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Zhao, G.; Zhang, S.; Li, Y.; Gu, H.; Li, Y.; Zhao, Q.; Qi, Y. Chloroplast-to-Nucleus Signaling Regulates MicroRNA Biogenesis in Arabidopsis. Dev. Cell 2019, 48, 371–382.e4. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.-R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, B.E.; Rivero, L.; Calhoun, C.S.; Grotewold, E.; Brkljacic, J. Standardized Method for High-throughput Sterilization of Arabidopsis Seeds. J. Vis. Exp. JoVE 2017, 128, e56587. [Google Scholar] [CrossRef]
- Conn, S.J.; Hocking, B.; Dayod, M.; Xu, B.; Athman, A.; Henderson, S.; Aukett, L.; Conn, V.; Shearer, M.K.; Fuentes, S.; et al. Protocol: Optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 2013, 9, 4. [Google Scholar] [CrossRef]



| miRNA | Target ID | Target Name | Target Function | References |
|---|---|---|---|---|
| ath-miR160b | AT2G28350.1 | ARF10 | Response to auxin signaling | [49,50] |
| AT4G30080.1 | ARF16 | |||
| AT1G77850.1 | ARF17 | |||
| ath-miR169f-3p | AT3G59770.1 | SAC9 | Probable phosphoinositide phosphatase | [51] |
| ath-miR394a | AT5G36170.2 | HCF109 | Proper translation, stability and processing of polycistronic transcripts in chloroplasts | [52] |
| ath-miR8175 | AT2G36250.2 | FtsZ2-1 | Required for plastid division | [53] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anna, B.-B.; Grzegorz, B.; Marek, K.; Piotr, G.; Marcin, F. Exposure to High-Intensity Light Systemically Induces Micro-Transcriptomic Changes in Arabidopsis thaliana Roots. Int. J. Mol. Sci. 2019, 20, 5131. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205131
Anna B-B, Grzegorz B, Marek K, Piotr G, Marcin F. Exposure to High-Intensity Light Systemically Induces Micro-Transcriptomic Changes in Arabidopsis thaliana Roots. International Journal of Molecular Sciences. 2019; 20(20):5131. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205131
Chicago/Turabian StyleAnna, Barczak-Brzyżek, Brzyżek Grzegorz, Koter Marek, Gawroński Piotr, and Filipecki Marcin. 2019. "Exposure to High-Intensity Light Systemically Induces Micro-Transcriptomic Changes in Arabidopsis thaliana Roots" International Journal of Molecular Sciences 20, no. 20: 5131. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205131