Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications
Abstract
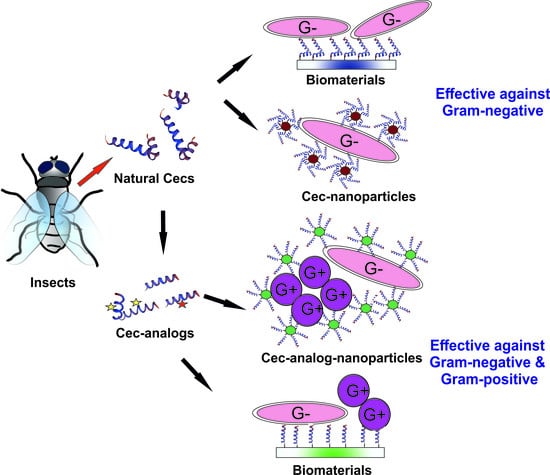
:1. Introduction
2. The Family of Cecropins in Insects
3. Cec Gene Expression and Mechanism of Action Against Microorganisms
4. In Vitro Antimicrobial Activity of Natural Cecs and Synthetic Cec-Analogs
5. Anti-Inflammatory Properties of Natural Cecs and Synthetic Cec-Analogs
6. Antitumor Activity of Natural Cecs and Synthetic Cec-Analogs
7. Health Benefits of Natural Cecs and Synthetic Cec-analogs: Future Potential and Limitations
7.1. Potential of Natural Cecs and Cec-analogs as Antibacterial Drugs
7.2. Natural Cecs and Cec-Analogs as Anti-Biofilm Compounds
7.3. Biomedical Applications of Natural Cecs and Cec-Analogs: Limitations and Potential Solutions
8. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Report to the Secretary General of the Nations: No Time to Wait–Securing the Future from Drug-Resistant Infections; Interagency Coordination Group on Antimicrobial Resistance (IACG): New York, NY, USA, 2019.
- Zhang, L.-J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14-9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2015, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Patočka, J.; Kuča, K. Insect antimicrobial peptides, a mini review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Tonk, M.; Vilcinskas, A. The medical potential of antimicrobial peptides from insects. Curr. Top. Med. Chem. 2017, 17, 554–575. [Google Scholar] [CrossRef]
- Yi, H.-Y.; Chowdhury, M.; Huang, Y.-D.; Yu, X.-Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef]
- Hultmark, D.; Steiner, H.; Rasmuson, T.; Boman, H.G. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur. J. Biochem. 1980, 106, 7–16. [Google Scholar] [CrossRef]
- Hultmark, D.; Engström, Å.; Bennich, H.; Kapur, R.; Boman, H.G. Insect immunity: Isolation and structure of cecropin D and four minor antibacterial components from Cecropia pupae. Eur. J. Biochem. 1982, 127, 207–217. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engström, Å.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246. [Google Scholar] [CrossRef]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150290. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Zhao, C.; Liaw, L.; Lee, I.H.; Lehrer, R.I. cDNA cloning of three cecropin-like antimicrobial peptides (Styelins) from the tunicate, Styela clava. FEBS Lett. 1997, 412, 144–148. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Boman, A.; Sun, C.X.; Andersson, M.; Jörnvall, H.; Mutt, V.; Boman, H.G. Antibacterial peptides from pig intestine: Isolation of a mammalian cecropin. Proc. Natl. Acad. Sci. USA 1989, 86, 9159–9162. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Boman, A.; Boman, H.G. Ascaris nematodes from pig and human make three anti-bacterial peptides: Isolation of cecropin P1 and two ASABF peptides. Cell. Mol. Life Sci. 2003, 60, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Pütsep, K.; Normark, S.; Boman, H.G. The origin of cecropins; implications from synthetic peptides derived from ribosomal protein L1. FEBS Lett. 1999, 451, 249–252. [Google Scholar] [CrossRef]
- Tamang, D.G.; Saier, M.H., Jr. The cecropin superfamily of toxic peptides. J. Mol. Microbiol. Biotechnol. 2006, 11, 94–103. [Google Scholar] [CrossRef]
- Tarr, D.E.K. Distribution and characteristics of ABFs, cecropins, nemapores, and lysozymes in nematodes. Dev. Comp. Immunol. 2012, 36, 502–520. [Google Scholar] [CrossRef]
- Segovia, L.J.T.; Ramirez, G.A.T.; Arias, D.C.H.; Duran, J.D.R.; Bedoya, J.P.; Osorio, J.C.C. Identification and characterization of novel cecropins from the Oxysternon conspicillatum neotropic dung beetle. PLoS ONE 2017, 12, e0187914. [Google Scholar] [CrossRef]
- Saito, A.; Ueda, K.; Imamura, M.; Atsumi, S.; Tabunoki, H.; Miura, N.; Watanabe, A.; Kitami, M.; Sato, R. Purification and cDNA cloning of a cecropin from the longicorn beetle, Acalolepta luxuriosa. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 142, 317–323. [Google Scholar] [CrossRef]
- Memarpoor-Yazdi, M.; Zare-Zardini, H.; Asoodeh, A. A novel antimicrobial peptide derived from the insect Paederus dermatitis. Int. J. Pept. Res. Ther. 2013, 19, 99–108. [Google Scholar] [CrossRef]
- Wu, J.; Mu, L.; Zhuang, L.; Han, Y.; Liu, T.; Li, J.; Yang, Y.; Yang, H.; Wei, L. A cecropin-like antimicrobial peptide with anti-inflammatory activity from the black fly salivary glands. Parasites Vectors 2015, 8, 561. [Google Scholar] [CrossRef] [PubMed]
- Vizioli, J.; Bulet, P.; Charlet, M.; Lowenberger, C.; Blass, C.; Müller, H.M.; Dimopoulos, G.; Hoffmann, J.; Kafatos, F.C.; Richman, A. Cloning and analysis of a cecropin gene from the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 2000, 9, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Lowenberger, C.; Charlet, M.; Vizioli, J.; Kamal, S.; Richman, A.; Christensen, B.M.; Bulet, P. Antimicrobial activity spectrum, cDNA cloning, and mRNA expression of a newly isolated member of the cecropin family from the mosquito vector Aedes aegypti. J. Biol. Chem. 1999, 274, 20092–20097. [Google Scholar] [CrossRef] [PubMed]
- Jayamani, E.; Rajamuthiah, R.; Larkins-Ford, J.; Fuchs, B.B.; Conery, A.L.; Vilcinskas, A.; Ausubel, F.M.; Mylonakis, E. Insect-derived cecropins display activity against Acinetobacter baumannii in a whole-animal high-throughput Caenorhabditis elegans model. Antimicrob. Agents Chemother. 2015, 59, 1728–1737. [Google Scholar] [CrossRef]
- Wei, L.; Yang, Y.; Zhou, Y.; Li, M.; Yang, H.; Mu, L.; Qian, Q.; Wu, J.; Xu, W. Anti-inflammatory activities of Aedes aegypti cecropins and their protection against murine endotoxin shock. Parasites Vectors 2018, 11, 470. [Google Scholar] [CrossRef]
- Sun, D.; Eccleston, E.D.; Fallon, A.M. Peptide sequence of an antibiotic cecropin from the vector mosquito, Aedes albopictus. Biochem. Biophys. Res. Commun. 1998, 249, 410–415. [Google Scholar] [CrossRef]
- Kaushal, A.; Gupta, K.; Shah, R.; van Hoek, M.L. Antimicrobial activity of mosquito cecropin peptides against Francisella. Dev. Comp. Immunol. 2016, 63, 171–180. [Google Scholar] [CrossRef]
- Xu, X.; Yang, H.; Ma, D.; Wu, J.; Wang, Y.; Song, Y.; Wang, X.; Lu, Y.; Yang, J.; Lai, R. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol. Cell. Proteomics 2008, 7, 582–590. [Google Scholar] [CrossRef]
- Park, S.I.; Yoe, S.M. A novel cecropin-like peptide from black soldier fly, Hermetia illucens: Isolation, structural and functional characterization. Entomol. Res. 2017, 47, 115–124. [Google Scholar] [CrossRef]
- Ekengren, S.; Hultmark, D. Drosophila cecropin as an antifungal agent. Insect Biochem. Mol. Biol. 1999, 29, 965–972. [Google Scholar] [CrossRef]
- Samakovlis, C.; Kimbrell, D.A.; Kylsten, P.; Engström, Å.; Hultmark, D. The immune response in Drosophila: Pattern of cecropin expression and biological activity. EMBO J. 1990, 9, 2969–2976. [Google Scholar] [CrossRef]
- Lu, X.; Shen, J.; Jin, X.; Ma, Y.; Huang, Y.; Mei, H.; Chu, F.; Zhu, J. Bactericidal activity of Musca domestica cecropin (Mdc) on multidrug-resistant clinical isolate of Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 95, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.M.; Jin, X.B.; Zhu, J.Y.; Mei, H.F.; Ma, Y.; Chu, F.J.; Wang, Y.; Li, X.B. Expression of the antimicrobial peptide cecropin fused with human lysozyme in Escherichia coli. Appl. Microbiol. Biotechnol. 2010, 87, 2169–2176. [Google Scholar] [CrossRef] [PubMed]
- Gui, S.; Li, R.; Feng, Y.; Wang, S. Transmission electron microscopic morphological study and flow cytometric viability assessment of Acinetobacter baumannii susceptible to Musca domestica cecropin. Sci. World J. 2014, 2014, 657536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulanger, N.; Brun, R.; Ehret-Sabatier, L.; Kunz, C.; Bulet, P. Immunopeptides in the defense reactions of Glossina morsitans to bacterial and Trypanosoma brucei brucei infections. Insect Biochem. Mol. Biol. 2002, 32, 369–375. [Google Scholar] [CrossRef]
- Boulanger, N.; Munks, R.J.; Hamilton, J.V.; Vovelle, F.; Brun, R.; Lehane, M.J.; Bulet, P. Epithelial innate immunity. A novel antimicrobial peptide with antiparasitic activity in the blood-sucking insect Stomoxys calcitrans. J. Biol. Chem. 2002, 277, 49921–49926. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Natori, S. Purification and characterization of an antibacterial protein from haemolymph of Sarcophaga peregrina (flesh-fly) larvae. Biochem. J. 1983, 211, 727–734. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Natori, S. Mode of action of a bactericidal protein induced in the haemolymph of Sarcophaga peregrina (flesh-fly) larvae. Biochem. J. 1984, 222, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Natori, S. Primary structure of sarcotoxin I, an antibacterial protein induced in the hemolymph of Sarcophaga peregrina (flesh fly) larvae. J. Biol. Chem. 1985, 260, 7174–7177. [Google Scholar]
- Pöppel, A.K.; Vogel, H.; Wiesner, J.; Vilcinskas, A. Antimicrobial peptides expressed in medicinal maggots of the blow fly Lucilia sericata show combinatorial activity against bacteria. Antimicrob. Agents Chemother. 2015, 59, 2508–2514. [Google Scholar] [CrossRef] [Green Version]
- De Lucca, A.J.; Bland, J.M.; Jacks, T.J.; Grimm, C.; Walsh, T.J. Fungicidal and binding properties of the natural peptides cecropin B and dermaseptin. Med. Mycol. 1998, 36, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Wachinger, M.; Kleinschmidt, A.; Winder, D.; von Pechmann, N.; Ludvigsen, A.; Neumann, M.; Holle, R.; Salmons, B.; Erfle, V.; Brack-Werner, R. Antimicrobial peptides melittin and cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J. Gen. Virol. 1998, 79, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Andrä, J.; Berninghausen, O.; Leippe, M. Cecropins, antibacterial peptides from insects and mammals, are potently fungicidal against Candida albicans. Med. Microbiol. Immunol. 2001, 189, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Shin, A.; Kim, Y. Anti-inflammatory activities of cecropin a and its mechanism of action. Arch. Insect Biochem. Physiol. 2015, 88, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.J.; Beazley, W.D.; Bibby, M.C.; Devine, D.A. Antimicrobial activity of cecropins. J. Antimicrob. Chemother. 1996, 37, 1077–1089. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Guo, C.; Huang, Y.; Zhang, X.; Chen, Y. Inhibition of porcine reproductive and respiratory syndrome virus by Cecropin D in vitro. Infect. Genet. Evol. 2015, 34, 7–16. [Google Scholar] [CrossRef]
- Qu, Z.; Steiner, H.; Engström, A.; Bennich, H.; Boman, H.G. Insect immunity: Isolation and structure of cecropins B and D from pupae of the Chinese oak silk moth, Antheraea pernyi. Eur. J. Biochem. 1982, 127, 219–224. [Google Scholar] [CrossRef]
- Wang, J.; Ma, K.; Ruan, M.; Wang, Y.; Li, Y.; Fu, Y.V.; Song, Y.; Sun, H.; Wang, J. A novel cecropin B-derived peptide with antibacterial and potential anti-inflammatory properties. PeerJ 2018, 6, e5369. [Google Scholar] [CrossRef]
- Fang, S.L.; Wang, L.; Fang, Q.; Chen, C.; Zhao, X.S.; Qian, C.; Wei, G.Q.; Zhu, B.J.; Liu, C.L. Characterization and functional study of a Cecropin-like peptide from the Chinese oak silkworm, Antheraea pernyi. Arch. Insect Biochem. Physiol. 2017, 94, e21368. [Google Scholar] [CrossRef]
- Yang, W.; Cheng, T.; Ye, M.; Deng, X.; Yi, H.; Huang, Y.; Tan, X.; Han, D.; Wang, B.; Xiang, Z.; et al. Functional divergence among silkworm antimicrobial peptide paralogs by the activities of recombinant proteins and the induced expression profiles. PLoS ONE 2011, 6, e18109. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Geng, T.; Hou, C.; Huang, Y.; Qin, G.; Guo, X. Bombyx mori cecropin A has a high antifungal activity to entomopathogenic fungus Beauveria bassiana. Gene 2016, 583, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Romoli, O.; Mukherjee, S.; Mohid, S.A.; Dutta, A.; Montali, A.; Franzolin, E.; Brady, D.; Zito, F.; Bergantino, E.; Rampazzo, C.; et al. Enhanced Silkworm Cecropin B Antimicrobial Activity against Pseudomonas aeruginosa from Single Amino Acid Variation. ACS Infect. Dis. 2019, 5, 1200–1213. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Mraheil, M.A.; Silva, S.; Müller, D.; Cemic, F.; Hemberger, J.; Hain, T.; Vilcinskas, A.; Chakraborty, T. Anti-Listeria activities of Galleria mellonella hemolymph proteins. Appl. Environ. Microbiol. 2011, 77, 4237–4240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oñate-Garzón, J.; Manrique-Moreno, M.; Trier, S.; Leidy, C.; Torres, R.; Patiño, E. Antimicrobial activity and interactions of cationic peptides derived from Galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. (Tokyo) 2017, 70, 238–245. [Google Scholar] [CrossRef]
- Kim, S.R.; Hong, M.Y.; Park, S.W.; Choi, K.H.; Yun, E.Y.; Goo, T.W.; Kang, S.W.; Suh, H.J.; Kim, I.; Hwang, J.S. Characterization and cDNA cloning of a cecropin-like antimicrobial peptide, papiliocin, from the swallowtail butterfly, Papilio xuthus. Mol. Cells 2010, 29, 419–423. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, E.; Shin, S.; Jeong, K.W.; Lee, J.Y.; Bae, S.Y.; Kim, S.H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. J. Biol. Chem. 2011, 286, 41296–41311. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Kim, J.-K.; Jeon, D.; Jeong, K.-W.; Shin, A.; Kim, Y. Functional roles of aromatic residues and helices of papiliocin in its antimicrobial and anti-inflammatory activities. Sci. Rep. 2015, 5, 12048. [Google Scholar] [CrossRef] [Green Version]
- Choi, C.-S.; Yoe, S.-M.; Kim, E.-S.; Chae, K.-S.; Kim, H.R. Purification and Characterization of Antibacterial Peptides, Spodopsin Ia and Ib Induced in the Larval Haemolymph of the Common Cutworm, Spodoptera Iitura. Korean J. Biol. Sci. 1997, 1, 457–462. [Google Scholar]
- Choi, C.S.; Lee, I.H.; Kim, E.; Kim, S.I.; Kim, H.R. Antibacterial properties and partial cDNA sequences of cecropin-like antibacterial peptides from the common cutworm, Spodoptera litura. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2000, 125, 287–297. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Du, C.; Chen, W.; Pang, Y. Characterization and expression of a cecropin-like gene from Helicoverpa armigera. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007, 148, 417–425. [Google Scholar] [CrossRef]
- Lockey, T.D.; Ourth, D.D. Formation of pores in Escherichia coli cell membranes by a cecropin isolated from hemolymph of Heliothis virescens larvae. Eur. J. Biochem. 1996, 236, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.R.; Lee, Y.H.; Bang, I.S.; Kim, E.S.; Kang, C.S.; Yun, C.Y.; Lee, I.H. cDNA cloning and antibacterial activities of cecropin D-like peptides from Agrius convolvuli. Arch. Insect Biochem. Physiol. 2000, 45, 149–155. [Google Scholar] [CrossRef]
- Park, S.I.; An, H.S.; Chang, B.S.; Yoe, S.M. Expression, cDNA cloning, and characterization of the antibacterial peptide cecropin D from Agrius convolvuli. Anim. Cells Syst. 2013, 17, 23–30. [Google Scholar] [CrossRef]
- Bang, I.S.; Son, S.Y.; Yoe, S.M. Hinnavin I, an antibacterial peptide from cabbage butterfly, Artogeia rapae. Mol. Cells 1997, 7, 509–513. [Google Scholar]
- Yoe, S.M.; Kang, C.S.; Han, S.S.; Bang, I.S. Characterization and cDNA cloning of hinnavin II, a cecropin family antibacterial peptide from the cabbage butterfly, Artogeia rapae. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006, 144, 199–205. [Google Scholar] [CrossRef]
- Duwadi, D.; Shrestha, A.; Yilma, B.; Kozlovski, I.; Sa-Eed, M.; Dahal, N.; Jukosky, J. Identification and screening of potent antimicrobial peptides in arthropod genomes. Peptides 2018, 103, 26–30. [Google Scholar] [CrossRef]
- Kylsten, P.; Samakovlis, C.; Hultmark, D. The cecropin locus in Drosophila; a compact gene cluster involved in the response to infection. EMBO J. 1990, 9, 217–224. [Google Scholar] [CrossRef]
- Tryselius, Y.; Samakovlis, C.; Kimbrell, D.A.; Hultmark, D. CecC, a cecropin gene expressed during metamorphosis in Drosophila pupae. Eur. J. Biochem. 1992, 204, 395–399. [Google Scholar] [CrossRef]
- Sackton, T.B.; Lazzaro, B.P.; Clark, A.G. Rapid expansion of immune-related gene families in the house fly, Musca domestica. Mol. Biol. Evol. 2017, 34, 857–872. [Google Scholar] [CrossRef] [Green Version]
- Gudmundsson, G.H.; Lidholm, D.A.; Asling, B.; Gan, R.; Boman, H.G. The cecropin locus. Cloning and expression of a gene cluster encoding three antibacterial peptides in Hyalophora cecropia. J. Biol. Chem. 1991, 266, 11510–11517. [Google Scholar]
- Ponnuvel, K.M.; Subhasri, N.; Sirigineedi, S.; Murthy, G.N.; Vijayaprakash, N.B. Molecular evolution of the cecropin multigene family in silkworm Bombyx mori. Bioinformation 2010, 5, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Ntwasa, M.; Goto, A.; Kurata, S. Coleopteran antimicrobial peptides: Prospects for clinical applications. Int. J. Microbiol. 2012, 2012, 101989. [Google Scholar] [CrossRef] [Green Version]
- Zou, Z.; Evans, J.D.; Lu, Z.; Zhao, P.; Williams, M.; Sumathipala, N.; Hetru, C.; Hultmark, D.; Jiang, H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007, 8, R177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quesada, H.; Ramos-Onsins, S.E.; Aguadé, M. Birth-and-death evolution of the Cecropin multigene family in Drosophila. J. Mol. Evol. 2005, 60, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tassanakajon, A.; Somboonwiwat, K.; Amparyup, P. Sequence diversity and evolution of antimicrobial peptides in invertebrates. Dev. Comp. Immunol. 2015, 48, 324–341. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Onsins, S.; Aguade, M. Molecular evolution of the cecropin multigene family in Drosophila: Functional genes vs. pseudogenes. Genetics 1998, 150, 157–171. [Google Scholar]
- Unckless, R.L.; Lazzaro, B.P. The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Phil. Tran. R. Soc. B 2016, 371, 20150291. [Google Scholar] [CrossRef] [Green Version]
- Tennessen, J.A. Molecular evolution of animal antimicrobial peptides: Widespread moderate positive selection. J. Evol. Biol. 2005, 18, 1387–1394. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Xia, L.-J.; Li, J.-Y.; Zhang, F.-C. CecropinXJ inhibits the proliferation of human gastric cancer BGC823 cells and induces cell death in vitro and in vivo. Int. J. Oncol. 2015, 46, 2181–2193. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Uvell, H.; Engström, Y. A multilayered defense against infection: Combinatorial control of insect immune genes. Trends Genet. 2007, 23, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Romoli, O.; Saviane, A.; Bozzato, A.; D’Antona, P.; Tettamanti, G.; Squartini, A.; Cappellozza, S.; Sandrelli, F. Differential sensitivity to infections and antimicrobial peptide-mediated immune response in four silkworm strains with different geographical origin. Sci. Rep. 2017, 7, 1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efimova, S.S.; Schagina, L.V.; Ostroumova, O.S. Channel-forming activity of cecropins in lipid bilayers: Effect of agents modifying the membrane dipole potential. Langmuir 2014, 30, 7884–7892. [Google Scholar] [CrossRef]
- Efimova, S.S.; Medvedev, R.Y.; Chulkov, E.G.; Schagina, L.V.; Ostroumova, O.S. Regulation of the Pore-Forming Activity of Cecropin A by Local Anesthetics. Cell Tiss. Biol. 2018, 12, 331–341. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [Green Version]
- Andreu, D.; Merrifield, R.B.; Steiner, H.; Boman, H.G. N-terminal analogs of cecropin A: Synthesis, antibacterial activity, and conformational properties. Biochemistry 1985, 24, 1683–1688. [Google Scholar] [CrossRef]
- Christensen, B.; Fink, J.; Merrifield, R.B.; Mauzerall, D. Channel-forming properties of cecropins and related model compounds incorporated into planar lipid membranes. Proc. Natl. Acad. Sci. USA 1988, 85, 5072–5076. [Google Scholar] [CrossRef] [Green Version]
- Durell, S.R.; Raghunathan, G.; Guy, H.R. Modeling the ion channel structure of cecropin. Biophys. J. 1992, 63, 1623–1631. [Google Scholar] [CrossRef] [Green Version]
- Tacconelli, E.; Magrini, N.; Kahlmeter, G.; Singh, N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; Volume 27. [Google Scholar]
- Jaynes, J.M.; Burton, C.A.; Barr, S.B.; Jeffers, G.W.; Julian, G.R.; White, K.L.; Enright, F.M.; Klei, T.R.; Laine, R.A. In vitro cytocidal effect of novel lytic peptides on Plasmodium falciparum and Trypanosoma cruzi. FASEB J. 1988, 2, 2878–2883. [Google Scholar] [CrossRef] [Green Version]
- De Lucca, A.J.; Bland, J.M.; Vigo, C.B.; Jacks, T.J.; Peter, J.; Walsh, T.J. D-cecropin B: Proteolytic resistance, lethality for pathogenic fungi and binding properties. Med. Mycol. 2000, 38, 301–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boman, H.G.; Wade, D.; Boman, I.A.; Wåhlin, B.; Merrifield, R.B. Antibacterial and antimalarial properties of peptides that are cecropin-melittin hybrids. FEBS Lett. 1989, 259, 103–106. [Google Scholar] [CrossRef] [Green Version]
- Saugar, J.M.; Rodríguez-Hernández, M.J.; de la Torre, B.G.; Pachón-Ibañez, M.E.; Fernández-Reyes, M.; Andreu, D.; Pachón, J.; Rivas, L. Activity of cecropin A-melittin hybrid peptides against colistin-resistant clinical strains of Acinetobacter baumannii: Molecular basis for the differential mechanisms of action. Antimicrob. Agents Chemother. 2006, 50, 1251–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garbacz, K.; Kamysz, W.; Piechowicz, L. Activity of antimicrobial peptides, alone or combined with conventional antibiotics, against Staphylococcus aureus isolated from the airways of cystic fibrosis patients. Virulence 2017, 8, 94–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geitani, R.; Moubareck, C.A.; Touqui, L.; Sarkis, D.K. Cationic antimicrobial peptides: Alternatives and/or adjuvants to antibiotics active against methicillin-resistant Staphylococcus aureus and multidrug-resistant Pseudomonas aeruginosa. BMC Microbiol. 2019, 19, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, S.; Li, W.; Zhang, L.; Zhang, Y.; Cao, B. Cecropin A–melittin mutant with improved proteolytic stability and enhanced antimicrobial activity against bacteria and fungi associated with gastroenteritis in vitro. Biochem. Biophys. Res. Commun. 2014, 451, 650–655. [Google Scholar] [CrossRef]
- Chicharro, C.; Granata, C.; Lozano, R.; Andreu, D.; Rivas, L. N-terminal fatty acid substitution increases the leishmanicidal activity of CA (1-7) M (2-9), a cecropin-melittin hybrid peptide. Antimicrob. Agents Chemother. 2001, 45, 2441–2449. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Lee, M.K.; Kim, K.L.; Hahm, K.S. Structure-antitumor and hemolytic activity relationships of synthetic peptides derived from cecropin A-magainin 2 and cecropin A-melittin hybrid peptides. J. Pept. Res. 1997, 50, 279–285. [Google Scholar] [CrossRef]
- Oh, D.; Shin, S.Y.; Lee, S.; Kang, J.H.; Kim, S.D.; Ryu, P.D.; Hahm, K.S.; Kim, Y. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A (1–8)–magainin 2 (1–12) and its analogues, on their antibiotic activities and structures. Biochemistry 2000, 39, 11855–11864. [Google Scholar] [CrossRef]
- Jeong, K.-W.; Shin, S.-Y.; Kim, J.-K.; Kim, Y.-M. Antibacterial activity and synergism of the hybrid antimicrobial peptide, CAMA-syn. Bull. Korean Chem. Soc. 2009, 30, 1839–1844. [Google Scholar]
- Ryu, S.; Choi, S.Y.; Acharya, S.; Chun, Y.J.; Gurley, C.; Park, Y.; Armstrong, C.A.; Song, P.I.; Kim, B.J. Antimicrobial and anti-inflammatory effects of Cecropin A(1–8)-Magainin2(1–12) hybrid peptide analog p5 against Malassezia furfur infection in human keratinocytes. J. Invest. Dermatol. 2011, 131, 1677–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.G.; Park, Y.; Jin, I.; Hahm, K.S.; Lee, H.H.; Moon, Y.H.; Woo, E.R. Structure-antiviral activity relationships of cecropin A-magainin 2 hybrid peptide and its analogues. J. Pept. Sci. 2004, 10, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Seo, C.H.; Luchian, T.; Park, Y. Antimicrobial Peptide CMA3 Derived from the CA-MA Hybrid Peptide: Antibacterial and Anti-inflammatory Activities with Low Cytotoxicity and Mechanism of Action in Escherichia coli. Antimicrob. Agents Chemother. 2015, 60, 495–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.-B.; Wu, R.-J.; Si, D.-Y.; Liao, X.-D.; Zhang, L.-L.; Zhang, R.-J. Novel hybrid peptide cecropin A (1–8)-LL37 (17–30) with potential antibacterial activity. Int. J. Mol. Sci. 2016, 17, 983. [Google Scholar] [CrossRef]
- Liu, D.; Liu, J.; Li, J.; Xia, L.; Yang, J.; Sun, S.; Ma, J.; Zhang, F. A potential food biopreservative, CecXJ-37N, non-covalently intercalates into the nucleotides of bacterial genomic DNA beyond membrane attack. Food Chem. 2017, 217, 576–584. [Google Scholar] [CrossRef]
- Wei, L.; Huang, C.; Yang, H.; Li, M.; Yang, J.; Qiao, X.; Mu, L.; Xiong, F.; Wu, J.; Xu, W. A potent anti-inflammatory peptide from the salivary glands of horsefly. Parasites Vectors. 2015, 8, 556. [Google Scholar] [CrossRef] [Green Version]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 2017, 7, 2173. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef]
- Kell, D.B.; Pretorius, E. On the translocation of bacteria and their lipopolysaccharides between blood and peripheral locations in chronic, inflammatory diseases: The central roles of LPS and LPS-induced cell death. Integr. Biol. (Camb.) 2015, 7, 1339–1377. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Jacob, B.; Jang, M.; Kwak, C.; Lee, Y.; Son, K.; Lee, S.; Jung, I.D.; Jeong, M.S.; Kwon, S.H.; et al. Development of a novel short 12-meric papiliocin-derived peptide that is effective against Gram-negative sepsis. Sci. Rep. 2019, 9, 3817. [Google Scholar] [CrossRef] [Green Version]
- Giacometti, A.; Cirioni, O.; Ghiselli, R.; Viticchi, C.; Mocchegiani, F.; Riva, A.; Saba, V.; Scalise, G. Effect of mono-dose intraperitoneal cecropins in experimental septic shock. Crit. Care Med. 2001, 29, 1666–1669. [Google Scholar] [CrossRef]
- Zhang, L.; Gui, S.; Liang, Z.; Liu, A.; Chen, Z.; Tang, Y.; Xiao, M.; Chu, F.; Liu, W.; Jin, X.; et al. Musca domestica Cecropin (Mdc) Alleviates Salmonella typhimurium-Induced Colonic Mucosal Barrier Impairment: Associating With Inflammatory and Oxidative Stress Response, Tight Junction as Well as Intestinal Flora. Front. Microbiol. 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.M.; Wang, W.; Smith, D.; Chan, S.C. Effects of the anti-bacterial peptide cecropin B and its analogs, cecropins B-1 and B-2, on liposomes, bacteria, and cancer cells. Biochim. Biophys. Acta 1997, 1336, 171–179. [Google Scholar] [CrossRef]
- Moore, A.J.; Devine, D.A.; Bibby, M.C. Preliminary experimental anticancer activity of cecropins. Pept. Res. 1994, 7, 265–269. [Google Scholar]
- Anghel, R.; Jitaru, D.; Bădescu, L.; Bădescu, M.; Ciocoiu, M. The cytotoxic effect of magainin II on the MDA-MB-231 and M14K tumour cell lines. Biomed. Res. Int. 2013, 2013, 831709. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.-C.; Hui, L.; Chen, H.M. Enhancement of the cytolytic effect of anti-bacterial cecropin by the microvilli of cancer cells. Anticancer Res. 1998, 18, 4467–4474. [Google Scholar]
- Jin, X.; Mei, H.; Li, X.; Ma, Y.; Zeng, A.H.; Wang, Y.; Lu, X.; Chu, F.; Wu, Q.; Zhu, J. Apoptosis-inducing activity of the antimicrobial peptide cecropin of Musca domestica in human hepatocellular carcinoma cell line BEL-7402 and the possible mechanism. Acta Biochim. Biophys. Sin. 2010, 42, 259–265. [Google Scholar] [CrossRef] [Green Version]
- Suttmann, H.; Retz, M.; Paulsen, F.; Harder, J.; Zwergel, U.; Kamradt, J.; Wullich, B.; Unteregger, G.; Stöckle, M.; Lehmann, J. Antimicrobial peptides of the Cecropin-family show potent antitumor activity against bladder cancer cells. BMC Urol. 2008, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Hilchie, A.L.; Hoskin, D.W.; Coombs, M.R.P. Anticancer Activities of Natural and Synthetic Peptides. In Antimicrobial Peptides. Advances in Experimental Medicine and Biology; Matsuzaki, K., Ed.; Springer: Singapore, 2019; Volume 1117, pp. 131–147. [Google Scholar] [CrossRef]
- Xia, L.J.; Wu, Y.L.; Ma, J.; Zhang, F.C. Therapeutic effects of antimicrobial peptide on malignant ascites in a mouse model. Mol. Med. Rep. 2018, 17, 6245–6252. [Google Scholar] [CrossRef] [Green Version]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jozefiak, A.; Engberg, R.M. Insect proteins as a potential source of antimicrobial peptides in livestock production. A review. J. Anim. Feed Sci. 2017, 26, 87–99. [Google Scholar] [CrossRef]
- Chiou, P.P.; Chen, M.J.; Lin, C.-M.; Khoo, J.; Larson, J.; Holt, R.; Leong, J.A.; Thorgarrd, G.; Chen, T.T. Production of homozygous transgenic rainbow trout with enhanced disease resistance. Mar. Biotechnol. 2014, 16, 299–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bundó, M.; Montesinos, L.; Izquierdo, E.; Campo, S.; Mieulet, D.; Guiderdoni, E.; Rossignol, M.; Badosa, E.; Montesinos, E.; San Segundo, B.; et al. Production of cecropin A antimicrobial peptide in rice seed endosperm. BMC Plant Biol. 2014, 14, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajesh, K.S.; Zareena, S.H.; Kumar, M.S.A. Assessment of salivary calcium, phosphate, magnesium, pH, and flow rate in healthy subjects, periodontitis, and dental caries. Contemp. Clin. Dent. 2015, 6, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Joris, L.; Dab, I.; Quinton, P.M. Elemental composition of human airway surface fluid in healthy and diseased airways. Am. Rev. Respir. Dis. 1993, 148, 1633–1637. [Google Scholar] [CrossRef]
- Shrestha, A.; Duwadi, D.; Jukosky, J.; Fiering, S.N. Cecropin-like antimicrobial peptide protects mice from lethal E. coli infection. PLoS ONE 2019, 14, e0220344. [Google Scholar] [CrossRef]
- Lewies, A.; Du Plessis, L.H.; Wentzel, J.F. Antimicrobial Peptides: The Achilles’ Heel of Antibiotic Resistance? Probiotics Antimicrob. Proteins 2019, 11, 370–381. [Google Scholar] [CrossRef]
- Rodríguez-Rojas, A.; Makarova, O.; Rolff, J. Antimicrobials, stress and mutagenesis. PLoS Pathog. 2014, 10, e1004445. [Google Scholar] [CrossRef] [Green Version]
- Grassi, L.; Maisetta, G.; Esin, S.; Batoni, G. Combination strategies to enhance the efficacy of antimicrobial peptides against bacterial biofilms. Front. Microbiol. 2017, 8, 2409. [Google Scholar] [CrossRef]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Dostert, M.; Belanger, C.R.; Hancock, R.E.W. Design and assessment of anti-biofilm peptides: Steps toward clinical application. J. Innate Immun. 2019, 11, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Riool, M.; de Breij, A.; Drijfhout, J.W.; Nibbering, P.H.; Zaat, S.A.J. Antimicrobial peptides in biomedical device manufacturing. Front. Chem. 2017, 5, 63. [Google Scholar] [CrossRef]
- Mataraci, E.; Dosler, S. In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 2012, 56, 6366–6371. [Google Scholar] [CrossRef] [Green Version]
- Cole, M.A.; Scott, T.F.; Mello, C.M. Bactericidal Hydrogels via Surface Functionalization with Cecropin A. ACS Biomater. Sci. Eng. 2016, 2, 1894–1904. [Google Scholar] [CrossRef]
- Querido, M.M.; Felgueiras, H.P.; Rai, A.; Costa, F.; Monteiro, C.; Borges, I.; Oliveira, D.; Ferreira, L.; Martins, M.C. Cecropin–Melittin Functionalized Polyurethane Surfaces Prevent Staphylococcus epidermidis Adhesion without Inducing Platelet Adhesion and Activation. Adv. Mater. Interfaces 2018, 5, 1801390. [Google Scholar] [CrossRef]
- Bai, L.; Zhu, L.; Min, S.; Liu, L.; Cai, Y.; Yao, J. Surface modification and properties of Bombyx mori silk fibroin films by antimicrobial peptide. Appl. Surf. Sci. 2008, 254, 2988–2995. [Google Scholar] [CrossRef]
- Saviane, A.; Romoli, O.; Bozzato, A.; Freddi, G.; Cappelletti, C.; Rosini, E.; Cappellozza, S.; Tettamanti, G.; Sandrelli, F. Intrinsic antimicrobial properties of silk spun by genetically modified silkworm strains. Transgenic Res. 2018, 27, 87–101. [Google Scholar] [CrossRef]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.; Dias, S.C.; Franco, O.L. Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef] [Green Version]
- Rai, A.; Pinto, S.; Velho, T.R.; Ferreira, A.F.; Moita, C.; Trivedi, U.; Evangelista, M.; Comune, M.; Rumbaugh, K.P.; Simões, P.N.; et al. One-step synthesis of high-density peptide-conjugated gold nanoparticles with antimicrobial efficacy in a systemic infection model. Biomaterials 2016, 85, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Bédard, F.; Biron, E. Recent progress in the chemical synthesis of class II and S-glycosylated bacteriocins. Front. Microbiol. 2018, 9, 1048. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, K.E.; Joski, P.; Johnston, K.J. Antibiotic-resistant infection treatment costs have doubled since 2002, now exceeding $2 billion annually. Health Affairs 2018, 37, 662–669. [Google Scholar] [CrossRef] [PubMed]


| Insect | Species | Active Peptide (aa) | Antimicrobial Activity | Peptide conc. (μM) | |||
|---|---|---|---|---|---|---|---|
| Order | Virus | Bacteria | Fungi | Cytotox. | Hem Act. | ||
| Coleoptera | Oxysternon conspicillatum | Oxysterlin 1 (39) [19] | - | G+, G− | weak | >28 | >14 |
| Oxysterlin 2 (55) [19] | - | G− | NA | >19.75 | >19.75 | ||
| Oxysterlin 3 (39) [19] | - | G− | NA | >28 | >28 | ||
| Acalolepta luxuriosa | Cec (35) [20] | - | M. luteus, E. coli | - | - | - | |
| Paederus dermatitis | Sarcotoxin Pd (34) [21] | - | G+, G− | weak | - | 16 | |
| Diptera | Simulium bannaense | SibaCec (35) [22] | - | G+, G− | - | 58 | 58 |
| Anopheles gambiae | AngCec A (35) [23] | - | G+, G− | A | - | - | |
| Aedes aegypti | AeaeCec 1 (34) [24,25,26] | - | G+, G− | A | 50 [26] | 50 [26] | |
| AeaeCec 2–4 (34) [26] | - | - | - | 50 | 50 | ||
| AeaeCec 5 (34) [26] | - | - | - | 12.5 | 12.5 | ||
| Aedes albopictus | Cec A1 (35) [27,28] | - | E. coli, Francisella | - | - | - | |
| Cec B (35) [28] | - | Francisella | - | - | - | ||
| Culex pipens | Cec A (34) [28] | - | Francisella | - | - | - | |
| Cec B2 (34) [28] | - | Francisella | - | - | - | ||
| Tabanus yao | Cec TY1 (41) [29] | - | B. subtilis S. aureus E. coli | A | - | - | |
| Hermetia illucens | CLP1 (45) [30] | - | G− | - | - | - | |
| Drosophila melanogaster | Cec A (34) [23,24,31,32] | - | G+, G− | A | - | - | |
| Cec B (34) [31,32] | - | G− | A | - | - | ||
| Musca domestica | Mdc (40) [33,34,35] | - | G+, G− | - | - | - | |
| Glossina morsitans | Cec (39) [36] | - | M. luteus, E. coli | - | - | - | |
| Stomoxys calcitrans | Stomoxyn (42) [37] | - | G+, G− | A | - | >10 | |
| Sarcophaga peregrina | Sarcotoxins I A, B, C (39) [38,39,40] | - | G+, G− | - | - | - | |
| Lucilia sericata | Lser Cecs 1–6 (40) [41] | - | G− | NA | - | - | |
| LSerStomox1 (43) [41] | - | G− | NA | - | - | ||
| LSerStomox 2 (42) [41] | - | G− | NA | - | - | ||
| Lepidoptera | Hyalophora cecropia | Cec A (37) [8,9,10,42,43,44,45] | HIV | G+, G− | A | [44,45] | 100 [45] |
| Cec B (35) [9,42,44,46] | - | G+, G− | A | 30 [44] | 500 [46] | ||
| Cec D (36) [9,47] | PRRSV | G+, G− | - | - | - | ||
| Antheraea pernyi | Cec B (35) [48,49] | - | G+, G− | 25 [49] | 200 [49] | ||
| Cec D (36) [48] | - | G+, G− | - | - | - | ||
| ApCec (38) [50] | - | B. subtilis, E. coli | - | - | 62.5 | ||
| Bombyx mori | Cec A (35) [51,52] | - | G+, G− | A | - | - | |
| Cec B (35) [51,53] | - | G+, G− | NA | 200 [53] | 200 [53] | ||
| Cec D (36) [51] | - | G+, G− | - | - | - | ||
| Cec E (?) [51] | - | B. thuringiensis, G− | - | - | - | ||
| Galleria mellonella | Cec D (39) [54,55] | - | L. monocytogenes | - | - | >115 [55] | |
| Papilio xuthus | Papiliocin (38) [56,57,58] | - | G+, G− | A | 12.5 [58] | 100 [58] | |
| Spodoptera litura | Spodopsin Ia (35) [59] | - | G+, G− | NA | - | - | |
| Spodopsin Ib (35) [59] | - | G+, G− | NA | - | - | ||
| Cec A (35) [60] | - | G+, G− | - | - | - | ||
| Cec B (35) [60] | - | G+, G− | - | - | - | ||
| Helicoverpa armigera | Cec D (42) [61] | - | G+, G− | - | - | - | |
| Heliothis virescens | Cec B (35) [62] | - | E. coli | - | - | - | |
| Agrius convolvuli | AcCec D 1-3 (38) [63,64] | - | G+, G− | - | - | - | |
| Artogeia rapa | Hinnavin I (40) [65] | - | G+, G− | A | - | - | |
| (Pieris rapae) | Hinnavin II (38) [66] | - | G+, G− | A | - | - | |
| Danaus plexippus | DAN1 (37) [67] | - | G+ (weak), G− | - | - | 49.56 | |
| DAN2 (37) [67] | - | G+ (weak), G− | weak | - | 48.97 | ||
| Peptide (aa) | Source | Modification | Antimicrobial Activity | Peptide Conc. (μM) | ||||
|---|---|---|---|---|---|---|---|---|
| Virus | Bacteria | Fungi | Protozoa | Cytotox. | Hem Act. | |||
| SB-37 (38) [92] | H. cecropia Cec B | aa add./sub. | - | - | - | P. falciparum, T. cruzi | - | - |
| Shiva-1 (38) [46,92] | H. cecropia Cec B | aa add./sub. | - | G+, G− | NA | P. falciparum, T. cruzi | - | - |
| D-Cec B (35) [93] | A. pernyi Cec B | D-enantiomer | - | - | A | - | - | - |
| CecDH (32) [49] | A. pernyi Cec B | aa del. | - | G+, G− | - | - | 25 | 100 |
| ΔM1 (39) [55] | G melonella Cec D | N-term aa sub. | - | Sa (weak), Ec, Pa | - | - | - | 115 |
| ΔM2 (39) [55] | G melonella Cec D | N-term aa sub. | - | Sa, Ec, Pa | - | - | - | ~60 |
| Mdc–hly (?) [34] | M. domestica Mdc; human Lysozyme | Hybrid | - | G+, G− | - | - | - | - |
| CAMs (≤26) [94,95,96,97,98] | H. cecropia Cec A; A. mellifera Mellitin | Hybrids | - | G+, G− | A | Plasmodium | 9 [96] | [98] |
| Ac-CAMs (15) [99] | H. cecropia Cec A; A. mellifera Mellitin | N-term fatty acid acylation | - | Sa, Ec, Ab | - | L. pifanoi | - | - |
| CAM-W (26) [98] | H. cecropia Cec A; A. mellifera Mellitin | aa sub. | - | G+, G− | A | - | - | 3.12 |
| CA-MAs (≤20) [100,101,102,103,104,105] | H-cecropia Cec A; X. laevis Magainin 2 | Hybrids with aa sub. | virus–cell fusion inhibition | G+, G− | A | - | [105] | [105] |
| CA-LL37 (22) [106] | H-cecropia Cec A; human LL37 | Hybrid | - | G+, G− | - | - | - | [106] |
| CecXJ-37C (37) [107] | B. mori Cec B | C-term aa add. | - | G+, G− | - | - | 20 | 19 |
| CecXJ-37N (37) [107] | B. mori Cec B | C-term aa add. | - | G+, G− | - | - | 20 | 33 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brady, D.; Grapputo, A.; Romoli, O.; Sandrelli, F. Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications. Int. J. Mol. Sci. 2019, 20, 5862. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20235862
Brady D, Grapputo A, Romoli O, Sandrelli F. Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications. International Journal of Molecular Sciences. 2019; 20(23):5862. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20235862
Chicago/Turabian StyleBrady, Daniel, Alessandro Grapputo, Ottavia Romoli, and Federica Sandrelli. 2019. "Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications" International Journal of Molecular Sciences 20, no. 23: 5862. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20235862