Deciphering miRNAs’ Action through miRNA Editing
Abstract
:1. Introduction
2. The Complex World of miRNA Biology—From Biogenesis to Action
3. Editing of miRNA
3.1. ADAR
3.2. AID/APOBEC
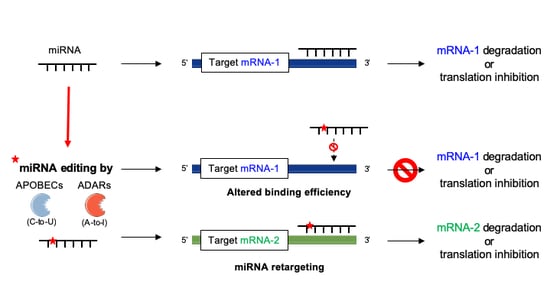
4. Editing in miRNA Biogenesis and Activity
4.1. miRNA Maturation
4.2. miRNA–mRNA Interactions
4.3. miRNA–ceRNA Interactions
4.4. Regulation of the miRNA Editing Machinery by RNA-Binding Proteins (RBPs)
5. Tools to Study miRNA Editing
6. miRNA-Dependent Regulation of Editing—Closing the Loop
7. miRNA Editing in Pathophysiological Processes
7.1. Development
7.2. Obesity and Metabolic Diseases
7.3. Inflammation and Immunity
7.4. Cancer
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigita, G.; Acunzo, M.; Romano, G.; Veneziano, D.; Lagana, A.; Vitiello, M.; Wernicke, D.; Ferro, A.; Croce, C.M. microRNA editing in seed region aligns with cellular changes in hypoxic conditions. Nucleic Acids Res. 2016, 44, 6298–6308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turunen, T.A.; Roberts, T.C.; Laitinen, P.; Vaananen, M.A.; Korhonen, P.; Malm, T.; Yla-Herttuala, S.; Turunen, M.P. Changes in nuclear and cytoplasmic microRNA distribution in response to hypoxic stress. Sci. Rep. 2019, 9, 10332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, A.; Mols, J.; Han, J. RISC-target interaction: Cleavage and translational suppression. Biochim. Biophys. Acta 2008, 1779, 668–677. [Google Scholar] [CrossRef] [Green Version]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar] [CrossRef] [Green Version]
- Hagan, J.P.; Piskounova, E.; Gregory, R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009, 16, 1021–1025. [Google Scholar] [CrossRef] [Green Version]
- Heo, I.; Joo, C.; Kim, Y.K.; Ha, M.; Yoon, M.J.; Cho, J.; Yeom, K.H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [Green Version]
- Thornton, J.E.; Chang, H.M.; Piskounova, E.; Gregory, R.I. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875–1885. [Google Scholar] [CrossRef] [Green Version]
- Nussbacher, J.K.; Yeo, G.W. Systematic Discovery of RNA Binding Proteins that Regulate MicroRNA Levels. Mol. Cell 2018, 69, 1005–1016.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Huang, T.; Zhang, X. RNA editing of microRNA prevents RNA-induced silencing complex recognition of target mRNA. Open Biol. 2015, 5, 150126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [Green Version]
- Sobolewski, C.; Calo, N.; Portius, D.; Foti, M. MicroRNAs in fatty liver disease. Semin. Liver Dis. 2015, 35, 12–25. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [Green Version]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef]
- Gjorgjieva, M.; Sobolewski, C.; Dolicka, D.; Correia de Sousa, M.; Foti, M. miRNAs and NAFLD: From pathophysiology to therapy. Gut 2019. [Google Scholar] [CrossRef]
- Leung, A.K.L. The Whereabouts of microRNA Actions: Cytoplasm and Beyond. Trends Cell Biol. 2015, 25, 601–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar, C.; Thum, T.; de Gonzalo-Calvo, D. Circulating miRNAs as mediators in cell-to-cell communication. Epigenomics 2019, 11, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.Y.; Ma, L.M.; Guo, Y.H.; Zhang, Y.C.; Zhou, H.; Shao, P.; Chen, Y.Q.; Qu, L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3’ trailers. PLoS ONE 2010, 5, e10563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krutzfeldt, J.; Poy, M.N.; Stoffel, M. Strategies to determine the biological function of microRNAs. Nat. Genet. 2006, 38, S14–S19. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Davidson, N.O. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 594–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnan, A.; Iyer, L.M.; Holland, S.J.; Boehm, T.; Aravind, L. Diversification of AID/APOBEC-like deaminases in metazoa: Multiplicity of clades and widespread roles in immunity. Proc. Natl. Acad. Sci. USA 2018, 115, E3201–E3210. [Google Scholar] [CrossRef] [Green Version]
- Qiu, S.; Li, W.; Xiong, H.; Liu, D.; Bai, Y.; Wu, K.; Zhang, X.; Yang, H.; Ma, K.; Hou, Y.; et al. Single-cell RNA sequencing reveals dynamic changes in A-to-I RNA editome during early human embryogenesis. BMC Genom. 2016, 17, 766. [Google Scholar] [CrossRef] [Green Version]
- Hwang, T.; Park, C.K.; Leung, A.K.; Gao, Y.; Hyde, T.M.; Kleinman, J.E.; Rajpurohit, A.; Tao, R.; Shin, J.H.; Weinberger, D.R. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 2016, 19, 1093–1099. [Google Scholar] [CrossRef]
- Sharma, S.; Patnaik, S.K.; Thomas Taggart, R.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015, 6, 6881. [Google Scholar] [CrossRef]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, E.C.; Washburn, M.C.; Major, F.; Rusch, D.B.; Hundley, H.A. Noncoding regions of C. elegans mRNA undergo selective adenosine to inosine deamination and contain a small number of editing sites per transcript. RNA Biol. 2015, 12, 162–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liang, H. When MicroRNAs Meet RNA Editing in Cancer: A Nucleotide Change Can Make a Difference. Bioessays 2018, 40. [Google Scholar] [CrossRef] [PubMed]
- Warnefors, M.; Liechti, A.; Halbert, J.; Valloton, D.; Kaessmann, H. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol. 2014, 15, R83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savva, Y.A.; Rieder, L.E.; Reenan, R.A. The ADAR protein family. Genome Biol. 2012, 13, 252. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.-D.; Öhman, M. ADAR1 Editing and its Role in Cancer. Genes 2018, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Melcher, T.; Maas, S.; Herb, A.; Sprengel, R.; Seeburg, P.H.; Higuchi, M. A mammalian RNA editing enzyme. Nature 1996, 379, 460–464. [Google Scholar] [CrossRef]
- Jain, M.; Mann, T.D.; Stulic, M.; Rao, S.P.; Kirsch, A.; Pullirsch, D.; Strobl, X.; Rath, C.; Reissig, L.; Moreth, K.; et al. RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure. EMBO J. 2018, 37, e94813. [Google Scholar] [CrossRef]
- Ishiguro, S.; Galipon, J.; Ishii, R.; Suzuki, Y.; Kondo, S.; Okada-Hatakeyama, M.; Tomita, M.; Ui-Tei, K. Base-pairing probability in the microRNA stem region affects the binding and editing specificity of human A-to-I editing enzymes ADAR1-p110 and ADAR2. RNA Biol. 2018, 15, 976–989. [Google Scholar] [CrossRef]
- Pestal, K.; Funk, C.C.; Snyder, J.M.; Price, N.D.; Treuting, P.M.; Stetson, D.B. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 2015, 43, 933–944. [Google Scholar] [CrossRef] [Green Version]
- Hartner, J.C.; Schmittwolf, C.; Kispert, A.; Muller, A.M.; Higuchi, M.; Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004, 279, 4894–4902. [Google Scholar] [CrossRef] [Green Version]
- Hartner, J.C.; Walkley, C.R.; Lu, J.; Orkin, S.H. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009, 10, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Deffit, S.N.; Hundley, H.A. To edit or not to edit: Regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 2016, 7, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Sinha, A.N.; Ray, A.; Lal, M.; Nayak, S.; Sharma, A.; Mehani, B.; Mukherjee, D.; Laddha, S.V.; Suri, A.; et al. A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Sci. Rep. 2017, 7, 2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [Green Version]
- Tomaselli, S.; Galeano, F.; Alon, S.; Raho, S.; Galardi, S.; Polito, V.A.; Presutti, C.; Vincenti, S.; Eisenberg, E.; Locatelli, F.; et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015, 16, 5. [Google Scholar] [CrossRef]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Baysal, B.E. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci. Rep. 2016, 6, 39100. [Google Scholar] [CrossRef]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef] [Green Version]
- Lerner, T.; Papavasiliou, F.N.; Pecori, R. RNA Editors, Cofactors, and mRNA Targets: An Overview of the C-to-U RNA Editing Machinery and Its Implication in Human Disease. Genes 2018, 10, 13. [Google Scholar] [CrossRef] [Green Version]
- Bennett, R.P.; Diner, E.; Sowden, M.P.; Lees, J.A.; Wedekind, J.E.; Smith, H.C. APOBEC-1 and AID are nucleo-cytoplasmic trafficking proteins but APOBEC3G cannot traffic. Biochem. Biophys. Res. Commun. 2006, 350, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, B.R.; Hamilton, C.E.; Mwangi, M.M.; Dewell, S.; Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 2011, 18, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Cole, D.C.; Chung, Y.; Gagnidze, K.; Hajdarovic, K.H.; Rayon-Estrada, V.; Harjanto, D.; Bigio, B.; Gal-Toth, J.; Milner, T.A.; McEwen, B.S.; et al. Loss of APOBEC1 RNA-editing function in microglia exacerbates age-related CNS pathophysiology. Proc. Natl. Acad. Sci. USA 2017, 114, 13272–13277. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Baysal, B.E. Stem-loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. PeerJ 2017, 5, e4136. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, X.; Huang, F.; Yang, B.; Li, J.; Liu, B.; Luo, H.; Zhang, P.; Zhang, H. APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. J. Biol. Chem. 2012, 287, 29373–29383. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Liang, Z.; Yang, B.; Tian, H.; Ma, J.; Zhang, H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J. Biol. Chem. 2007, 282, 33632–33640. [Google Scholar] [CrossRef] [Green Version]
- Ding, Q.; Chang, C.J.; Xie, X.; Xia, W.; Yang, J.Y.; Wang, S.C.; Wang, Y.; Xia, J.; Chen, L.; Cai, C.; et al. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J. Clin. Investig. 2011, 121, 4526–4536. [Google Scholar] [CrossRef]
- Blanc, V.; Park, E.; Schaefer, S.; Miller, M.; Lin, Y.; Kennedy, S.; Billing, A.M.; Ben Hamidane, H.; Graumann, J.; Mortazavi, A.; et al. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 2014, 15, R79. [Google Scholar] [CrossRef] [Green Version]
- Warf, M.B.; Shepherd, B.A.; Johnson, W.E.; Bass, B.L. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 2012, 22, 1488–1498. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; Megraw, M.; Kreider, E.; Iizasa, H.; Valente, L.; Hatzigeorgiou, A.G.; Nishikura, K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008, 36, 5270–5280. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007, 8, 763–769. [Google Scholar] [CrossRef]
- Li, L.; Song, Y.; Shi, X.; Liu, J.; Xiong, S.; Chen, W.; Fu, Q.; Huang, Z.; Gu, N.; Zhang, R. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2018, 28, 132–143. [Google Scholar] [CrossRef] [Green Version]
- Iizasa, H.; Wulff, B.E.; Alla, N.R.; Maragkakis, M.; Megraw, M.; Hatzigeorgiou, A.; Iwakiri, D.; Takada, K.; Wiedmer, A.; Showe, L.; et al. Editing of Epstein-Barr Virus-encoded BART6 MicroRNAs Controls Their Dicer Targeting and Consequently Affects Viral Latency. J. Biol. Chem. 2010, 285, 33358–33370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Lopez, J.; Hourcade Jde, D.; Del Mazo, J. Reprogramming of microRNAs by adenosine-to-inosine editing and the selective elimination of edited microRNA precursors in mouse oocytes and preimplantation embryos. Nucleic Acids Res. 2013, 41, 5483–5493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scadden, A.D. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005, 12, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Shibutani, T.; Nakanishi, N.; Nishikura, K.; Iwai, S.; Kuraoka, I. Human endonuclease V is a ribonuclease specific for inosine-containing RNA. Nat. Commun. 2013, 4, 2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallego, A.; Hartasanchez, D.A.; Braso-Vives, M.; Garcia-Ramallo, E.; Lopez-Valenzuela, M.; Baena, N.; Guitart, M.; Fernandez-Bellon, H.; Kondova, I.; Bontrop, R.; et al. RNA editing independently occurs at three mir-376a-1 sites and may compromise the stability of the microRNA hairpin. Gene 2017, 628, 109–116. [Google Scholar] [CrossRef]
- Cho, C.J.; Jung, J.; Jiang, L.; Lee, E.J.; Kim, D.S.; Kim, B.S.; Kim, H.S.; Jung, H.Y.; Song, H.J.; Hwang, S.W.; et al. Combinatory RNA-Sequencing Analyses Reveal a Dual Mode of Gene Regulation by ADAR1 in Gastric Cancer. Dig. Dis. Sci. 2018, 63, 1835–1850. [Google Scholar] [CrossRef]
- Chawla, G.; Sokol, N.S. ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res. 2014, 42, 5245–5255. [Google Scholar] [CrossRef] [Green Version]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015, 17, 311–321. [Google Scholar] [CrossRef]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.-T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef] [Green Version]
- Kume, H.; Hino, K.; Galipon, J.; Ui-Tei, K. A-to-I editing in the miRNA seed region regulates target mRNA selection and silencing efficiency. Nucleic Acids Res. 2014, 42, 10050–10060. [Google Scholar] [CrossRef] [Green Version]
- Gu, T.; Buaas, F.W.; Simons, A.K.; Ackert-Bicknell, C.L.; Braun, R.E.; Hibbs, M.A. Canonical A-to-I and C-to-U RNA Editing Is Enriched at 3′UTRs and microRNA Target Sites in Multiple Mouse Tissues. PLoS ONE 2012, 7, e33720. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hui, H.; Guo, Z.; Zhang, W.; Hu, Y.; He, T.; Tai, Y.; Peng, P.; Wang, L. ADAR1 regulates ARHGAP26 gene expression through RNA editing by disrupting miR-30b-3p and miR-573 binding. RNA 2013, 19, 1525–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yang, C.-S.; Varelas, X.; Monti, S. Altered RNA editing in 3’ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep. 2016, 6, 23226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, M.; Fukami, T.; Gotoh, S.; Takamiya, M.; Aoki, Y.; Nakajima, M. RNA Editing Modulates Human Hepatic Aryl Hydrocarbon Receptor Expression by Creating MicroRNA Recognition Sequence. J. Biol. Chem. 2016, 291, 894–903. [Google Scholar] [CrossRef] [Green Version]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar]
- Cai, Y.; Wan, J. Competing Endogenous RNA Regulations in Neurodegenerative Disorders: Current Challenges and Emerging Insights. Front. Mol. Neurosci. 2018, 11, 370. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Yan, Y.; Zeng, S.; Dai, S.; Chen, X.; Wei, J.; Gong, Z. Circular RNAs: Clinical relevance in cancer. Oncotarget 2018, 9, 1444–1460. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wang, J.; Khanabdali, R.; Kalionis, B.; Tai, X.; Xia, S. Circular RNAs: Isolation, characterization and their potential role in diseases. RNA Biol. 2017, 14, 1715–1721. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Ren, W.; Zhang, L.; Li, S.; Kong, X.; Zhang, H.; Dong, J.; Cai, G.; Jin, C.; Zheng, D.; et al. PTENp1, a natural sponge of miR-21, mediates PTEN expression to inhibit the proliferation of oral squamous cell carcinoma. Mol. Carcinog. 2017, 56, 1322–1334. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, Y.; Ma, Z.; Ma, G.; Xue, Q.; Li, F.; Liu, L. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. Oncotarget 2017, 8, 26079–26089. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Tang, X.; Su, L. Overexpression of Long Noncoding RNA PTENP1 Inhibits Cell Proliferation and Migration via Suppression of miR-19b in Breast Cancer Cells. Oncol. Res. 2018, 26, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Li, R.K.; Gao, J.; Guo, L.H.; Huang, G.Q.; Luo, W.H. PTENP1 acts as a ceRNA to regulate PTEN by sponging miR-19b and explores the biological role of PTENP1 in breast cancer. Cancer Gene Ther. 2017, 24, 309–315. [Google Scholar] [CrossRef]
- Nigita, G.; Marceca, G.P.; Tomasello, L.; Distefano, R.; Calore, F.; Veneziano, D.; Romano, G.; Nana-Sinkam, S.P.; Acunzo, M.; Croce, C.M. ncRNA Editing: Functional Characterization and Computational Resources. Methods Mol. Biol. 2019, 1912, 133–174. [Google Scholar] [PubMed]
- Mattick, J.S. The State of Long Non-Coding RNA Biology. Noncoding RNA 2018, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poria, D.K.; Guha, A.; Nandi, I.; Ray, P.S. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 2016, 35, 1703–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balkhi, M.Y.; Iwenofu, O.H.; Bakkar, N.; Ladner, K.J.; Chandler, D.S.; Houghton, P.J.; London, C.A.; Kraybill, W.; Perrotti, D.; Croce, C.M.; et al. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci. Signal. 2013, 6, ra63. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Karki, N.; Bhattacharya, C.; Zhu, R.; MacDuff, D.A.; Stenglein, M.D.; Schumacher, A.J.; Demorest, Z.L.; Harris, R.S.; Matin, A.; et al. APOBEC3 inhibits DEAD-END function to regulate microRNA activity. BMC Mol. Biol. 2013, 14, 16. [Google Scholar] [CrossRef] [Green Version]
- Wang, I.X.; So, E.; Devlin, J.L.; Zhao, Y.; Wu, M.; Cheung, V.G. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013, 5, 849–860. [Google Scholar] [CrossRef] [Green Version]
- Stellos, K.; Gatsiou, A.; Stamatelopoulos, K.; Perisic Matic, L.; John, D.; Lunella, F.F.; Jae, N.; Rossbach, O.; Amrhein, C.; Sigala, F.; et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016, 22, 1140–1150. [Google Scholar] [CrossRef]
- Anantharaman, A.; Tripathi, V.; Khan, A.; Yoon, J.H.; Singh, D.K.; Gholamalamdari, O.; Guang, S.; Ohlson, J.; Wahlstedt, H.; Ohman, M.; et al. ADAR2 regulates RNA stability by modifying access of decay-promoting RNA-binding proteins. Nucleic Acids Res. 2017, 45, 4189–4201. [Google Scholar] [CrossRef] [Green Version]
- Anantharaman, A.; Gholamalamdari, O.; Khan, A.; Yoon, J.H.; Jantsch, M.F.; Hartner, J.C.; Gorospe, M.; Prasanth, S.G.; Prasanth, K.V. RNA-editing enzymes ADAR1 and ADAR2 coordinately regulate the editing and expression of Ctn RNA. FEBS Lett. 2017, 591, 2890–2904. [Google Scholar] [CrossRef] [Green Version]
- Jinnah, H.; Ulbricht, R.J. Using mouse models to unlock the secrets of non-synonymous RNA editing. Methods 2019, 156, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, P.; Schneider, M.F.; Wettengel, J.; Stafforst, T. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew. Chem. Int. Ed. Engl. 2014, 53, 6267–6271. [Google Scholar] [CrossRef] [PubMed]
- Montiel-Gonzalez, M.F.; Vallecillo-Viejo, I.; Yudowski, G.A.; Rosenthal, J.J. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl. Acad. Sci. USA 2013, 110, 18285–18290. [Google Scholar] [CrossRef] [Green Version]
- Vallecillo-Viejo, I.C.; Liscovitch-Brauer, N.; Montiel-Gonzalez, M.F.; Eisenberg, E.; Rosenthal, J.J.C. Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol. 2018, 15, 104–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, P.; Moschref, M.; Li, Q.; Merkle, T.; Selvasaravanan, K.D.; Li, J.B.; Stafforst, T. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods 2018, 15, 535–538. [Google Scholar] [CrossRef]
- Guo, Y.; Yu, H.; Samuels, D.C.; Yue, W.; Ness, S.; Zhao, Y.Y. Single-nucleotide variants in human RNA: RNA editing and beyond. Brief Funct. Genom. 2019, 18, 30–39. [Google Scholar] [CrossRef]
- Giraldez, M.D.; Spengler, R.M.; Etheridge, A.; Godoy, P.M.; Barczak, A.J.; Srinivasan, S.; De Hoff, P.L.; Tanriverdi, K.; Courtright, A.; Lu, S.; et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018, 36, 746–757. [Google Scholar] [CrossRef]
- Ramaswami, G.; Li, J.B. Identification of human RNA editing sites: A historical perspective. Methods 2016, 107, 42–47. [Google Scholar] [CrossRef] [Green Version]
- Alon, S.; Mor, E.; Vigneault, F.; Church, G.M.; Locatelli, F.; Galeano, F.; Gallo, A.; Shomron, N.; Eisenberg, E. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012, 22, 1533–1540. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Baras, A.S.; Halushka, M.K. miRge 2.0 for comprehensive analysis of microRNA sequencing data. BMC Bioinform. 2018, 19, 275. [Google Scholar] [CrossRef]
- Maemura, K.; Watanabe, K.; Ando, T.; Hiyama, N.; Sakatani, T.; Amano, Y.; Kage, H.; Nakajima, J.; Yatomi, Y.; Nagase, T.; et al. Altered editing level of microRNAs is a potential biomarker in lung adenocarcinoma. Cancer Sci. 2018, 109, 3326–3335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vesely, C.; Tauber, S.; Sedlazeck, F.J.; von Haeseler, A.; Jantsch, M.F. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 2012, 22, 1468–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, K.; Auvinen, E.; Greco, D.; Auvinen, P. miRSeqNovel: An R based workflow for analyzing miRNA sequencing data. Mol. Cell. Probes 2012, 26, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Sablok, G.; Milev, I.; Minkov, G.; Minkov, I.; Varotto, C.; Yahubyan, G.; Baev, V. isomiRex: Web-based identification of microRNAs, isomiR variations and differential expression using next-generation sequencing datasets. FEBS Lett. 2013, 587, 2629–2634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, H.; Marzi, M.J.; Nicassio, F. IsomiRage: From Functional Classification to Differential Expression of miRNA Isoforms. Front. Bioeng. Biotechnol. 2014, 2, 38. [Google Scholar] [CrossRef] [Green Version]
- Kuenne, C.; Preussner, J.; Herzog, M.; Braun, T.; Looso, M. MIRPIPE: Quantification of microRNAs in niche model organisms. Bioinformatics 2014, 30, 3412–3413. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Dong, M.; Li, L.; Liu, L.; Luz-Madrigal, A.; Tsonis, P.A.; Del Rio-Tsonis, K.; Liang, C. mirPRo-a novel standalone program for differential expression and variation analysis of miRNAs. Sci. Rep. 2015, 5, 14617. [Google Scholar] [CrossRef] [Green Version]
- Pantano, L.; Estivill, X.; Marti, E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010, 38, e34. [Google Scholar] [CrossRef] [Green Version]
- Giurato, G.; De Filippo, M.R.; Rinaldi, A.; Hashim, A.; Nassa, G.; Ravo, M.; Rizzo, F.; Tarallo, R.; Weisz, A. iMir: An integrated pipeline for high-throughput analysis of small non-coding RNA data obtained by smallRNA-Seq. BMC Bioinform. 2013, 14, 362. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Batzel, P.; Sydes, J.; Eames, B.F.; Postlethwait, J.H. miRNA analysis with Prost! Reveals evolutionary conservation of organ-enriched expression and post-transcriptional modifications in three-spined stickleback and zebrafish. Sci. Rep. 2019, 9, 3913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, C.; Gao, J.; Zhang, H.; Jiang, X.; Zang, Q.; Ban, R.; Zhang, Y.; Shi, Q. CPSS 2.0: A computational platform update for the analysis of small RNA sequencing data. Bioinformatics 2017, 33, 3289–3291. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jang, I.; Jun, Y.; Yoon, S.; Ko, M.; Kwon, Y.; Choi, I.; Chang, H.; Ryu, D.; Lee, B.; et al. MiRGator v3.0: A microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013, 41, D252–D257. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liu, Q.; Wang, X.; Zheng, J.; Wang, T.; You, M.; Sheng Sun, Z.; Shi, Q. mirTools 2.0 for non-coding RNA discovery, profiling, and functional annotation based on high-throughput sequencing. RNA Biol. 2013, 10, 1087–1092. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zang, Q.; Zhang, H.; Ban, R.; Yang, Y.; Iqbal, F.; Li, A.; Shi, Q. DeAnnIso: A tool for online detection and annotation of isomiRs from small RNA sequencing data. Nucleic Acids Res. 2016, 44, W166–W175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aparicio-Puerta, E.; Lebron, R.; Rueda, A.; Gomez-Martin, C.; Giannoukakos, S.; Jaspez, D.; Medina, J.M.; Zubkovic, A.; Jurak, I.; Fromm, B.; et al. sRNAbench and sRNAtoolbox 2019: Intuitive fast small RNA profiling and differential expression. Nucleic Acids Res. 2019, 47, W530–W535. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Zheng, J.; Chen, Z.; Liu, Y.; Dura, B.; Kwak, M.; Xavier-Ferrucio, J.; Lu, Y.C.; Zhang, M.; Roden, C.; et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 2019, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef]
- Nemlich, Y.; Greenberg, E.; Ortenberg, R.; Besser, M.J.; Barshack, I.; Jacob-Hirsch, J.; Jacoby, E.; Eyal, E.; Rivkin, L.; Prieto, V.G.; et al. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J. Clin. Investig. 2013, 123, 2703–2718. [Google Scholar] [CrossRef] [Green Version]
- Caval, V.; Suspene, R.; Shapira, M.; Vartanian, J.P.; Wain-Hobson, S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage. Nat. Commun. 2014, 5, 5129. [Google Scholar] [CrossRef] [PubMed]
- Ekdahl, Y.; Farahani, H.S.; Behm, M.; Lagergren, J.; Ohman, M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 2012, 22, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Zhao, L.; Yang, L.; Huang, P.; Zhao, F.; Li, W.; Liu, Y. RNA editing by ADAR2 is metabolically regulated in pancreatic islets and beta-cells. J. Biol. Chem. 2006, 281, 33386–33394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Huang, P.; Li, F.; Zhao, L.; Zhang, Y.; Li, S.; Gan, Z.; Lin, A.; Li, W.; Liu, Y. c-Jun amino-terminal kinase-1 mediates glucose-responsive upregulation of the RNA editing enzyme ADAR2 in pancreatic beta-cells. PLoS ONE 2012, 7, e48611. [Google Scholar] [CrossRef]
- Singh, M.; Kesterson, R.A.; Jacobs, M.M.; Joers, J.M.; Gore, J.C.; Emeson, R.B. Hyperphagia-mediated obesity in transgenic mice misexpressing the RNA-editing enzyme ADAR2. J. Biol. Chem. 2007, 282, 22448–22459. [Google Scholar] [CrossRef] [Green Version]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715. [Google Scholar] [CrossRef] [Green Version]
- Zipeto, M.A.; Court, A.C.; Sadarangani, A.; Delos Santos, N.P.; Balaian, L.; Chun, H.-J.; Pineda, G.; Morris, S.R.; Mason, C.N.; Geron, I.; et al. ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell 2016, 19, 177–191. [Google Scholar] [CrossRef]
- Liu, G.; Ma, X.; Wang, Z.; Wakae, K.; Yuan, Y.; He, Z.; Yoshiyama, H.; Iizasa, H.; Zhang, H.; Matsuda, M.; et al. Adenosine deaminase acting on RNA-1 (ADAR1) inhibits HBV replication by enhancing microRNA-122 processing. J. Biol. Chem. 2019, 294. [Google Scholar] [CrossRef]
- Sharma, S.; Wang, J.; Alqassim, E.; Portwood, S.; Cortes Gomez, E.; Maguire, O.; Basse, P.H.; Wang, E.S.; Segal, B.H.; Baysal, B.E. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019, 20, 37. [Google Scholar] [CrossRef] [Green Version]
- Forrest, A.R.R.; Kanamori-Katayama, M.; Tomaru, Y.; Lassmann, T.; Ninomiya, N.; Takahashi, Y.; de Hoon, M.J.L.; Kubosaki, A.; Kaiho, A.; Suzuki, M.; et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 2009, 24, 460. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Rajewsky, K. MicroRNA Control in the Immune System: Basic Principles. Cell 2009, 137, 380. [Google Scholar] [CrossRef] [Green Version]
- Kalla, R.; Ventham, N.T.; Kennedy, N.A.; Quintana, J.F.; Nimmo, E.R.; Buck, A.H.; Satsangi, J. MicroRNAs: New players in IBD. Gut 2015, 64, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eltzschig, H.K.; Carmeliet, P. Hypoxia and inflammation. N. Engl. J. Med. 2011, 364, 656–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tassinari, V.; Cesarini, V.; Silvestris, D.A.; Gallo, A. The adaptive potential of RNA editing-mediated miRNA-retargeting in cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Cesarini, V.; Silvestris, D.A.; Tassinari, V.; Tomaselli, S.; Alon, S.; Eisenberg, E.; Locatelli, F.; Gallo, A. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res. 2018, 46, 2045–2059. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20246249
Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ Action through miRNA Editing. International Journal of Molecular Sciences. 2019; 20(24):6249. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20246249
Chicago/Turabian StyleCorreia de Sousa, Marta, Monika Gjorgjieva, Dobrochna Dolicka, Cyril Sobolewski, and Michelangelo Foti. 2019. "Deciphering miRNAs’ Action through miRNA Editing" International Journal of Molecular Sciences 20, no. 24: 6249. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20246249