The Effect of Transcription Factor MYB14 on Defense Mechanisms in Vitis quinquangularis-Pingyi
Abstract
:1. Introduction
2. Results
2.1. R2R3-MYB Transcription Factors Involved in Plant Defense
2.2. Distribution of MYB14 in the Grapevine Chromosome and Sequence Analyses of VqMYB14
2.3. Overexpression of VqMYB14 Enhanced the Stilbene Contents and Expression of Stilbene Biosynthesis Genes
2.4. pVqMYB14 Activation is Stronger than pVvMYB14 Activation after flg22 and Harpin Induction
2.5. Differences in pVqMYB14 and pVvMYB14 Activities Depend on Structural Differences of the Promoters
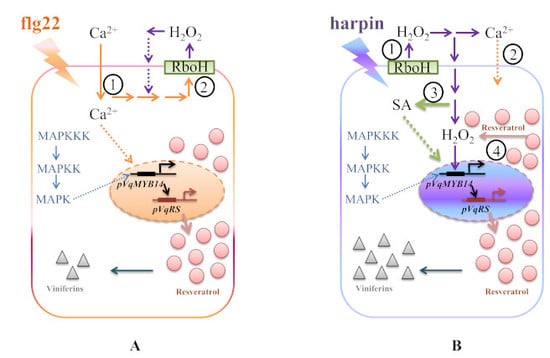
2.6. Early Upstream Signaling Events Involved in flg22- and Harpin-Triggered Immunity of pVqMYB14
2.7. The Role of SA in VqMYB14 Induction
2.8. flg22-Induced VqMYB14 Activation is More Sensitive to Gd Ions in Grapevine Than in a Heterologous Tobacco System
2.9. Harpin-Induced VqMYB14 Activation is More Sensitive to DPI in Grapevine Than in a Heterologous Tobacco System
2.10. The Harpin-Induced Oxidative Burst Occurs Earlier Than flg22-Induced
3. Discussion
3.1. Calcium Influx
3.2. Oxidative Burst
3.3. MAPK Signaling
3.4. SA Signaling
3.5. Stilbenes Output
4. Materials and Methods
4.1. Plant Materials
4.2. Gene Isolation and Sequence Analysis
4.3. Overexpression of VqMYB14 in V. quinquangularis-PY
4.4. Plasmid Construction
4.5. Transient Expression
4.6. Treatment of Tobacco Leaves and Grapevine Protoplasts for Transient Promoter Assays
4.7. Determination of H2O2 and Stilbene Levels in Grapevine Leaves
4.8. cDNA Synthesis and Quantitative Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nürnberger, T.; Lipka, V. Nonhost resistance in plants: New insights into an old phenomenon. Mol. Plant Pathol. 2005, 6, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Qiao, F.; Chang, X.L.; Nick, P. The cytoskeleton enhances gene expression in the response to the Harpin elicitor in grapevine. J. Exp. Bot. 2010, 61, 4021–4031. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.L.; Nick, P. Defence signalling triggered by Flg22 and Harpin is integrated into a different stilbene output in Vitis cells. PLoS ONE 2012, 7, e40446. [Google Scholar] [CrossRef]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef]
- Dong, H.; Delaney, T.P.; Bauer, D.W.; Beer, S.V. Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J. 1999, 20, 207–215. [Google Scholar] [CrossRef]
- Tampakaki, A.P.; Skandalis, N.A.; Gazi, D.; Bastaki, M.N.; Sarris, P.F.; Charova, S.N.; Kokkinidis, M.; Panopoulos, N.J. Playing the ‘‘Harp’’: Evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 2010, 48, 347–370. [Google Scholar] [CrossRef]
- Tsuda, K.; Sato, M.; Glazebrook, J.; Cohen, J.D.; Katagiri, F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008, 53, 763–775. [Google Scholar] [CrossRef]
- Katagiri, F.; Tsuda, K. Understanding the plant immune system. Mol. Plant Microbe Interact. 2010, 23, 1531–1536. [Google Scholar] [CrossRef] [Green Version]
- Chang, X.; Seo, M.; Takebayashi, Y.; Kamiya, Y.; Riemann, M.; Nick, P. Jasmonates are induced by the PAMP flg22 but not the cell deathinducing elicitor Harpin in Vitis rupestris. Protoplasma 2017, 254, 1–13. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Zeledon, J.; Zipper, R.; Spring, O. Assessment of phenotypic diversity of Plasmopara viticola on Vitis genotypes with different resistance. Crop Prot. 2013, 54, 221–228. [Google Scholar] [CrossRef]
- Rouxel, M.; Mestre, P.; Comont, G.; Lehman, B.L.; Schilder, A.; Delmotte, F. Phylogenetic and experimental evidence for host-specialized cryptic species in a biotrophic oomycete. New Phytol. 2013, 197, 251–263. [Google Scholar] [CrossRef]
- Gong, P.; Riemann, M.; Duan, D.; Nadja, S.; Bernadette, G.; Armin, M.; Nick, P. Two grapevine metacaspase genes mediate ETI-like cell death in grapevine defence against infection of Plasmopara viticola. Protoplasma 2019, 256, 951–969. [Google Scholar] [CrossRef]
- Tisch, C.; Nick, P.; Kortekamp, A. Rescue to be rescued: European wild grape as genetic resources of resistance towards fungal diseases. In Proceedings of the 7th International Workshop on Gapevine Downy and Powdery Mildew, Vitoria-Gasteiz, Spain, 30 June–4 July 2014; pp. 61–62, ISBN 978-84-7821-827-1. [Google Scholar]
- Qiu, W.; Feechan, A.; Dry, I. Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Hortic. Res. 2015, 2, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Schwaninger, H.; He, P.; Wang, Y. Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 2007, 46, 132–136. [Google Scholar]
- Pezet, R.; Gindro, K.; Viret, O.; Spring, J.L. Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol. Mol. Plant Pathol. 2004, 65, 297–303. [Google Scholar] [CrossRef]
- Jiao, Y.; Xu, W.; Duan, D.; Wang, Y.; Nick, P. A stilbene synthase allele from a Chinese wild grapevine confers resistance to powdery mildew by recruiting salicylic acid signalling for efficient defence. J. Exp. Bot. 2016, 67, 5841–5856. [Google Scholar] [CrossRef] [Green Version]
- Wong, D.C.J.; Schlechter, R.; Vannozzi, A.; Höll, J.; Hmmam, I.; Bogs, J.; Tornielli, G.B.; Castellarin, S.D.; Matus, J.T. A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 2016, 23, 451–466. [Google Scholar] [CrossRef]
- Wong, D.C.J.; Matus, J.T. Constructing integrated networks for identifying new secondary metabolic pathway regulators in grapevine: Recent applications and future opportunities. Front. Plant Sci. 2017, 8, 505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höll, J.; Vannozzi, A.; Czemmel, S.; D’Onofrio, C.; Walker, A.R.; Rausch, T.; Lucchin, M.; Boss, P.K.; Dry, I.B.; Bogs, J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 2013, 25, 4135–4149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Du, H.; Feng, B.R.; Yang, S.S.; Huang, Y.B.; Tang, Y.X. The R2R3-MYB Transcription Factor Gene Family in Maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef] [Green Version]
- Deluc, L.; Barrieu, F.; Marchive, C.; Lauvergeat, V.; Decendit, A.; Richard, T.; Carde, J.P.; Mérillon, J.M.; Hamdi, S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006, 140, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.-M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef] [Green Version]
- Bogs, J.; Jaffé, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.Z.; Wang, F.B.; Jin, C.; Tong, Y.; Wang, T. A R2R3-MYB transcription factor VvMYBF1 from grapevine (Vitis vinifera L.) regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis. J. Hortic. Sci. Biotechnol. 2019, 1–15. [Google Scholar] [CrossRef]
- Duan, D.; Fischer, S.; Merz, P.; Bogs, J.; Riemann, M.; Nick, P. An ancestral allele of grapevine transcription factor MYB14 promotes plant defence. J. Exp. Bot. 2016, 67, 1795–1804. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.Q.; Wang, L.; Zhu, J.L.; Chen, T.T.; Wang, Y.J.; Xu, Y. Histological responses to downy mildew in resistant and susceptible grapevines. Protoplasma 2015, 252, 259–270. [Google Scholar] [CrossRef]
- Yin, X.; Liu, R.Q.; Su, H. Pathogen development and host responses to Plasmopara viticola in resistant and susceptible grapevines: An ultrastructural study. Hortic. Res. 2017, 4, 17033. [Google Scholar] [CrossRef] [PubMed]
- Diaz-De-Leon, F.; Klotz, K.L.; Lagrimini, M. Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene. Plant Physiol. 1993, 101, 1117–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, K.; Numata, M.; Nakajima, I.; Goto-Yamamoto, N.; Matsumura, H.; Tanaka, N. Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J. Exp. Bot. 2014, 65, 4433–4449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buer, C.S.; Imin, N.; Djordjevic, M.A. Flavonoids: New Roles for Old Molecules. J. Integr. Plant Biol. 2010, 52, 100–113. [Google Scholar] [CrossRef]
- Pastuglia, M.; Roby, D.; Dumas, C.; Cock, J.M. Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase in Brassica oleracea. Plant Cell 1997, 9, 1–13. [Google Scholar]
- Zhu, Z.; Li, G.; Liu, L.; Zhang, Q.; Han, Z.; Chen, X.; Li, B. A R2R3-MYB Transcription Factor, VvMYBC2L2, Functions as a Transcriptional Repressor of Anthocyanin Biosynthesis in Grapevine (Vitis vinifera L.). Molecules 2018, 24, 92. [Google Scholar] [CrossRef] [Green Version]
- Vannozzi, A.; Wong, D.C.J.; Höll, J.; Hmmam, I.; Matus, J.T.; Bogs, J.; Ziegler, T.; Dry, I.; Barcaccia, G.; Lucchin, M. Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol. 2018, 59, 1043–1059. [Google Scholar] [CrossRef]
- Haas, H.U.; Alleweldt, G. The karyotype of grapevine (Vitis vinifera L.). Acta Hortic. 2000, 528, 249–258. [Google Scholar] [CrossRef]
- Xu, W.; Yu, Y.; Ding, J.; Hua, Z.; Wang, Y. Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 2010, 231, 475–487. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chow, C.N.; Zheng, H.Q.; Wu, N.Y.; Chien, C.H.; Huang, H.D.; Lee, T.Y.; Chiang-Hsieh, Y.F.; Hou, P.F.; Yang, T.Y.; Chang, W.C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabs, T.; Tschöpe, M.; Colling, C.; Hahlbrock, K.; Scheel, D. Elicitor stimulated ion fluxes and O2- from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. USA 1997, 94, 4800–4805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, J.P.; Pickard, B.G. Mechanosensory calcium-selective cation channels in epidermal cells. Plant J. 1993, 3, 83–110. [Google Scholar] [CrossRef] [PubMed]
- Wojtaszek, P. Oxidative burst: An early plant response to pathogen infection. Biochem. J. 1997, 322, 681–692. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.J.; Liu, Y.; He, P.; Lamikanra, O.; Lu, L. Resistance of Chinese Vitis species to Elsinoe ampelina (de Bary) Shear. Hortic. Sci. 1998, 33, 123–126. [Google Scholar]
- He, P.C. Viticulture; China Agriculture Press: Beijing, China, 1999. [Google Scholar]
- Liu, C.H. Study on Taxonomy and Geographical Distribution of Wild Vitis in China. Ph.D. Thesis, Henan Agricultural University, Zhengzhou, Henan, China, 2012. [Google Scholar]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signalling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Ohura, I.; Kawakita, K.; Yokota, N.; Fujiwara, M.; Shimamoto, K.; Yoshioka, N. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 2007, 19, 1065–1080. [Google Scholar] [CrossRef] [Green Version]
- Ogasawara, Y.; Kaya, H.; Hiraoka, G.; Yumoto, F.; Kimura, S.; Kadota, Y.; Hishinuma, H.; Senzaki, E.; Yamagoe, S.; Nagata, K.; et al. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 2008, 283, 8885–8892. [Google Scholar] [CrossRef] [Green Version]
- Giri, P.; Kumar, A.; Taj, G. In silico-prediction of downstream MYB interacting partners of MAPK3 in Arabidopsis. Bioinformation 2014, 10, 721–725. [Google Scholar] [CrossRef] [Green Version]
- Tenhaken, R.; Rübel, C. Salicylic acid is needed in hypersensitive cell death in soybean but does not act as a catalase inhibitor. Plant Physiol. 1997, 115, 291–298. [Google Scholar] [CrossRef] [Green Version]
- Adrian, M.; Jeandet, P.; Veneau, J.; Weston, L.A.; Bessis, R. Biological activity of resveratrol, a stilbenic compound from grapevines, against Botrytis cinerea, the causal agent for gray mold. J. Chem. Ecol. 1997, 23, 1689–1702. [Google Scholar] [CrossRef]
- Pezet, R.; Gindro, K.; Viret, O.; Richter, H. Effects of resveratrol, viniferins and pterostilbenes on Plasmopara viticola zoospore mobility and disease development. Vitis 2004, 43, 145–148. [Google Scholar]
- Chang, X.; Heene, E.; Qiao, F.; Nick, P. The Phytoalexin Resveratrol Regulates the Initiation of Hypersensitive Cell Death in Vitis Cell. PLoS ONE 2011, 6, e26405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mersereau, M.; Pazour, G.; Das, A. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 1990, 90, 149–151. [Google Scholar] [CrossRef]
- Gao, L.; Guo, X.; Liu, X.Q.; Zhang, L.; Huang, J.; Tan, L.; Lin, Z.; Nagawa, S.; Wang, D.Y. Changes in mitochondrial DNA levels during early embryogenesis in Torenia fournieri and Arabidopsis thaliana. Plant J. 2018, 95, 785–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, F.L.; Li, Y.J.; Hu, Y.; Gao, R.; Zang, X.W.; Ding, Q.; Wang, Y.J.; Wen, Y.Q. A highly efficient grapevine mesophyll protoplast system for transient gene expression and the study of disease resistance proteins. Plant Cell Tissue Organ Cult. 2016, 125, 43–57. [Google Scholar] [CrossRef]
- Pike, S.M.; Adam, A.L.; Pu, X.A.; Hoyos, M.E.; Laby, R.; Beer, S.V.; Novacky, A. Effects of Erwinia amylovora harpin on tobacco leaf cell membranes are related to leaf necrosis and electrolyte leakage and distinct from perturbations caused by inoculated E. amylovora. Physiol. Mol. Plant Pathol. 1998, 53, 39–60. [Google Scholar] [CrossRef]
- Duan, D.; Halter, D.; Baltenweck, R.; Tisch, C.; Tröster, V.; Kortekamp, A.; Hugueney, P.; Nick, P. Genetic diversity of stilbene metaboliam in Vitis sylvestris. J. Exp. Bot. 2015, 66, 3243–3257. [Google Scholar] [CrossRef]
- Zhang, M.X.; Li, Q.; Liu, T.L.; Liu, L.; Shen, D.Y.; Zhu, Y.; Liu, P.H.; Zhou, J.M.; Dou, D.L. Two cytoplasmic effectors of Phytophthora sojae regulate plant cell deah via interactions with plant catalases. Plant Physiol. 2015, 167, 164–175. [Google Scholar] [CrossRef] [Green Version]










© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Wang, Q.; Bai, R.; Li, R.; Chen, L.; Xu, Y.; Zhang, M.; Duan, D. The Effect of Transcription Factor MYB14 on Defense Mechanisms in Vitis quinquangularis-Pingyi. Int. J. Mol. Sci. 2020, 21, 706. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21030706
Luo Y, Wang Q, Bai R, Li R, Chen L, Xu Y, Zhang M, Duan D. The Effect of Transcription Factor MYB14 on Defense Mechanisms in Vitis quinquangularis-Pingyi. International Journal of Molecular Sciences. 2020; 21(3):706. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21030706
Chicago/Turabian StyleLuo, Yangyang, Qingyang Wang, Ru Bai, Ruixiang Li, Lu Chen, Yifan Xu, Ming Zhang, and Dong Duan. 2020. "The Effect of Transcription Factor MYB14 on Defense Mechanisms in Vitis quinquangularis-Pingyi" International Journal of Molecular Sciences 21, no. 3: 706. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21030706