Author Contributions
Conceptualization, N.F.; methodology, M.N. and N.F.; validation, G.G. and C.P.; formal analysis, G.L., M.N., C.C., and N.F.; investigation, G.L., C.C., G.G., E.S., and N.F.; resources, G.G., S.M., S.F., S.B., and N.F.; data curation, G.L., C.C., E.S., K.V., and D.G.; writing—original draft preparation, G.L. and C.C.; writing—review and editing, M.N., G.G., E.S., K.V., D.G., L.R., J.C., C.P., F.G., and N.F.; visualization, G.L., E.S., K.V., and N.F.; supervision, S.M., S.F., and S.B.; project administration, S.B. and N.F.; funding acquisition, S.F., S.B., and N.F. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Overview of 280 PTEN-low breast carcinomas. Heatmap illustrating the clinical, histologic, and biological information together with mismatch repair protein status of all PTEN low (i.e., scores 0 and 1) cases identified. Each column represents a case, each row a parameter, which is color-coded according to the legend below. ER, estrogen receptor; PR, progesterone receptor; MLH1, mutL homologue 1; MSH2, mutS homologue 2; MSH6, mutS homologue 6; PMS2, postmeiotic segregation increased 2 (PMS2); MMR, mismatch repair; LUM A, Luminal A; LUM B, Luminal B; TNBC, triple-negative breast cancer; IHC, immunohistochemistry.
Figure 1.
Overview of 280 PTEN-low breast carcinomas. Heatmap illustrating the clinical, histologic, and biological information together with mismatch repair protein status of all PTEN low (i.e., scores 0 and 1) cases identified. Each column represents a case, each row a parameter, which is color-coded according to the legend below. ER, estrogen receptor; PR, progesterone receptor; MLH1, mutL homologue 1; MSH2, mutS homologue 2; MSH6, mutS homologue 6; PMS2, postmeiotic segregation increased 2 (PMS2); MMR, mismatch repair; LUM A, Luminal A; LUM B, Luminal B; TNBC, triple-negative breast cancer; IHC, immunohistochemistry.
Figure 2.
PTEN and mismatch repair protein expression across different biomarker-based subgroups of breast cancer patients. ER, estrogen receptor; TNBC, triple-negative breast cancer; pMMR, mismatch repair proficient; hMMR, mismatch repair heterogeneous; dMMR, mismatch repair deficient.
Figure 2.
PTEN and mismatch repair protein expression across different biomarker-based subgroups of breast cancer patients. ER, estrogen receptor; TNBC, triple-negative breast cancer; pMMR, mismatch repair proficient; hMMR, mismatch repair heterogeneous; dMMR, mismatch repair deficient.
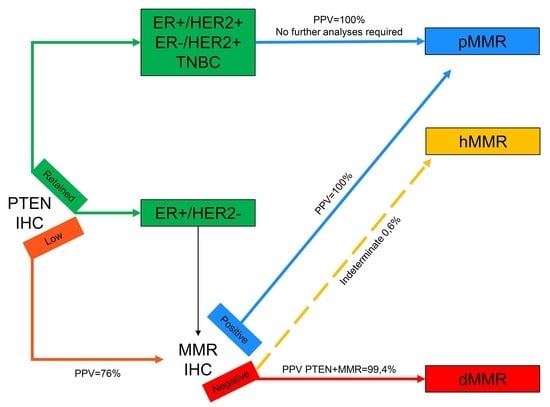
Figure 3.
Revised diagnostic algorithm for the evaluation of mismatch repair status in breast cancer. Connectors drawn with continuous lines represent links between different steps of the diagnostic workflow; connectors drawn with discontinuous lines represent indeterminate biological characteristics. All connectors are color-coded based on the different biological features represented in the squared boxes. IHC, immunohistochemistry; ER, estrogen receptor; TNBC, triple-negative breast cancer; PPV, positive predictive value; MMR, mismatch repair; pMMR, mismatch repair proficient; hMMR, mismatch repair heterogeneous; dMMR, mismatch repair deficient.
Figure 3.
Revised diagnostic algorithm for the evaluation of mismatch repair status in breast cancer. Connectors drawn with continuous lines represent links between different steps of the diagnostic workflow; connectors drawn with discontinuous lines represent indeterminate biological characteristics. All connectors are color-coded based on the different biological features represented in the squared boxes. IHC, immunohistochemistry; ER, estrogen receptor; TNBC, triple-negative breast cancer; PPV, positive predictive value; MMR, mismatch repair; pMMR, mismatch repair proficient; hMMR, mismatch repair heterogeneous; dMMR, mismatch repair deficient.
Figure 4.
Patterns of MMR protein expression. (A). Proficient status in the presence of retained nuclear expression. (B). Heterogeneous expression where only a subset of neoplastic nuclei is positive (arrows). (C) Deficiency of the expression across the entire tumor in the presence of internal positive controls (arrows). Example of MLH1 immunohistochemistry; original magnification: 400×.
Figure 4.
Patterns of MMR protein expression. (A). Proficient status in the presence of retained nuclear expression. (B). Heterogeneous expression where only a subset of neoplastic nuclei is positive (arrows). (C) Deficiency of the expression across the entire tumor in the presence of internal positive controls (arrows). Example of MLH1 immunohistochemistry; original magnification: 400×.
Figure 5.
Patterns of PTEN expression and scoring system employed in the present study. Score 0 was assigned in the absence of immunohistochemical staining in the tumor cells; score 1 was assigned when the neoplastic cells showed a weaker staining than the normal counterpart; score 2 was assigned in cases with equal staining intensity between tumor and normal epithelial/stromal cells; Cases were clustered as PTEN-low (score 1 and score 0) and PTEN-retained (score 2). Original magnification: 100×.
Figure 5.
Patterns of PTEN expression and scoring system employed in the present study. Score 0 was assigned in the absence of immunohistochemical staining in the tumor cells; score 1 was assigned when the neoplastic cells showed a weaker staining than the normal counterpart; score 2 was assigned in cases with equal staining intensity between tumor and normal epithelial/stromal cells; Cases were clustered as PTEN-low (score 1 and score 0) and PTEN-retained (score 2). Original magnification: 100×.
Table 1.
Correlation between phosphatase and tensin (PTEN) and mismatch repair status across selected clinicopathologic features. SD, standard deviation; ER, estrogen receptor; MMR, mismatch repair.
Table 1.
Correlation between phosphatase and tensin (PTEN) and mismatch repair status across selected clinicopathologic features. SD, standard deviation; ER, estrogen receptor; MMR, mismatch repair.
| | PTEN Low | PTEN Retained | p-Value |
|---|
| All patients, n (%) | 280 (46.1) | 328 (53.9) | |
| Age at diagnosis, mean ± SD | 61.5 ± 12.0 | 60.6 ± 13.6 | 0.3425 |
| Histological subtype, n (%) | | | 0.6243 |
| Ductal | 224 (80.0) | 267 (81.4) |
| Lobular | 40 (14.3) | 39 (11.9) |
| Other | 16 (5.7) | 22 (6.7) |
| Clinical cluster, n (%) | | | <0.0001 |
| ER+, HER2- | 176 (62.9) | 276 (84.2) |
| HER2+ | 63 (22.5) | 34 (10.4) |
| ER-, HER2- | 41 (14.6) | 18 (5.5) |
| Grade, n (%) | | | 0.6428 |
| 1 | 34 (12.1) | 39 (11.9) |
| 2 | 116 (41.4) | 148 (45.1) |
| 3 | 130 (46.4) | 141 (43.0) |
| T, n (%) | | | 0.7479 |
| 1 | 180 (64.3) | 198 (60.4) |
| 2 | 83 (29.6) | 105 (32.0) |
| 3 | 6 (2.1) | 8 (2.4) |
| 4 | 11 (3.9) | 17 (5.2) |
| N, n (%) | | | 0.2528 |
| - | 164 (58.6) | 207 (63.1) |
| + | 116 (41.4) | 121 (36.9) |
| Stage, n (%) | | | 0.4682 |
| 0, 1 | 118 (42.1) | 154 (47.0) |
| 2 | 107 (38.2) | 112 (34.2) |
| 3, 4 | 55 (19.6) | 62 (18.9) |
| MMR, n (%) | | | <0.0001 |
| Proficient | 174 (62.1) | 275 (83.8) |
| Deficient | 66 (23.6) | 15 (4.6) |
| Heterogeneous | 40 (14.3) | 38 (11.6) |
Table 2.
Bivariate analysis showing the association of selected clinicopathologic characteristics with patients’ death. ER, estrogen receptor; MMR, mismatch repair; MLH1, mutL homologue 1; MSH2, mutS homologue 2; MSH6, mutS homologue 6; PMS2, postmeiotic segregation increased 2 (PMS2).
Table 2.
Bivariate analysis showing the association of selected clinicopathologic characteristics with patients’ death. ER, estrogen receptor; MMR, mismatch repair; MLH1, mutL homologue 1; MSH2, mutS homologue 2; MSH6, mutS homologue 6; PMS2, postmeiotic segregation increased 2 (PMS2).
| | Death | p-Value |
|---|
| Yes | No |
|---|
| (n = 34) | (n = 569) |
|---|
| ER+/HER2, n (%) | | | 0.004 |
| ER+, HER2- | 17 (4.0) | 430 (96.0) |
| HER2+ | 11 (11.3) | 86 (88.7) |
| ER-, HER2- | 6 (10.2) | 53 (89.8) |
| Stage, n (%) | | | 0.0006 |
| 0, 1 | 8 (3.0) | 263 (97.0) |
| 2 | 12 (5.6) | 204 (94.4) |
| 3, 4 | 15 (12.9) | 101 (87.1) |
| PTEN retained, n (%) | 8 (2.5) | 317 (97.5) | 0.0001 |
| MMR, n (%) | | | 0.0161 |
| Proficient or heterogeneous | 25 (4.8) | 499 (95.2) |
| Deficient | 10 (12.7) | 69 (87.3) |
| MLH1, n (%) | | | 0.1712 |
| Proficient or heterogeneous | 30 (5.4) | 528 (94.6) |
| Deficient | 5 (11.2) | 40 (88.8) |
| MSH2, n (%) | | | 0.0107 |
| Proficient or heterogeneous | 27 (4.9) | 520 (95.1) |
| Deficient | 8 (14.3) | 48 (85.7) |
| MSH6, n (%) | | | 0.0641 |
| Proficient or heterogeneous | 31 (8.2) | 545 (91.8) |
| Deficient | 4 (14.8) | 23 (85.2) |
| PMS2, n (%) | | | 1 |
| Proficient or heterogeneous | 34 (5.8) | 549 (94.2) |
| Deficient | 1 (5.0) | 19 (95.0) |
Table 3.
Bivariate analysis showing the association of selected clinicopathologic characteristics with tumor progression in Luminal A breast cancers. MLH1, mutL homologue 1; MSH2, mutS homologue 2; MSH6, mutS homologue 6; PMS2, postmeiotic segregation increased 2 (PMS2); MMR, mismatch repair.
Table 3.
Bivariate analysis showing the association of selected clinicopathologic characteristics with tumor progression in Luminal A breast cancers. MLH1, mutL homologue 1; MSH2, mutS homologue 2; MSH6, mutS homologue 6; PMS2, postmeiotic segregation increased 2 (PMS2); MMR, mismatch repair.
| | Progression | p-Value |
|---|
| Yes | No |
|---|
| (n = 8) | (n = 150) |
|---|
| Stage, n (%) | | | 0.0346 |
| 0, 1 | 1 (1.3) | 79 (98.7) |
| 2 | 5 (7.6) | 61 (92.4) |
| 3, 4 | 2 (16.7) | 10 (83.3) |
| PTEN retained, n (%) | 1 (1.1) | 90 (98.9) | 0.0106 |
| MMR, n (%) | | | 0.3084 |
| Proficient or heterogeneous | 6 (4.4) | 130 (95.6) |
| Deficient | 2 (9.1) | 20 (89.9) |
| MLH1, n (%) | | | 0.4462 |
| Proficient or heterogeneous | 7 (4.8) | 140 (95.2) |
| Deficient | 1 (9.1) | 10 (90.9) |
| MSH2, n (%) | | | 0.1687 |
| Proficient or heterogeneous | 6 (4.2) | 137 (95.8) |
| Deficient | 2 (13.3) | 13 (86.7) |
| MSH6, n (%) | | | 0.0416 |
| Proficient or heterogeneous | 6 (4.0) | 145 (96.0) |
| Deficient | 2 (25.0) | 6 (75.0) |
| PMS2, n (%) | | | 1 |
| Proficient or heterogeneous | 8 (5.2) | 147 (94.8) |
| Deficient | 0 | 3 (100) |
Table 4.
Clinicopathologic factors associated with tumor progression in Luminal B breast cancers. Model defined using a stepwise selection of the predictors; significance levels for entry (SLE = 0.25) and for stay (SLS = 0.20). NST, invasive carcinoma of no special type (ductal); HR, hazard ratio; CI, confidence interval.
Table 4.
Clinicopathologic factors associated with tumor progression in Luminal B breast cancers. Model defined using a stepwise selection of the predictors; significance levels for entry (SLE = 0.25) and for stay (SLS = 0.20). NST, invasive carcinoma of no special type (ductal); HR, hazard ratio; CI, confidence interval.
| | HR | 95% CI | p-Value |
|---|
| Histological subtype, NST vs. other | 0.39 | 0.14-1.06 | 0.0653 |
| Systemic metastases | 11.9 | 2.01–71.1 | 0.0064 |
| Stage | | | |
| 2 vs. (0, 1) | 1.94 | 0.69-5.47 | 0.2377 |
| (3, 4) vs. (0, 1) | 3.56 | 1.29-9.81 | 0.0055 |
| PTEN-low | 3.24 | 1.37-7.65 | 0.0073 |
| Lymphovascular invasion | 2.31 | 0.85-6.29 | 0.1002 |
Table 5.
Multivariable analysis showing the association of selected clinicopathologic characteristics and PTEN expression with MMR status. NST, invasive carcinoma of no special type (ductal); ILC, invasive lobular carcinoma; TNBC, triple-negative breast cancer; OR, odds ratio; CI, confidence interval; dMMR, mismatch repair-deficient; pMMR, mismatch repair proficient; hMMR, mismatch repair heterogeneous.
Table 5.
Multivariable analysis showing the association of selected clinicopathologic characteristics and PTEN expression with MMR status. NST, invasive carcinoma of no special type (ductal); ILC, invasive lobular carcinoma; TNBC, triple-negative breast cancer; OR, odds ratio; CI, confidence interval; dMMR, mismatch repair-deficient; pMMR, mismatch repair proficient; hMMR, mismatch repair heterogeneous.
| | dMMR vs. pMMR | hMMR vs. pMMR |
|---|
| OR | 95% CI | p-value | OR | 95% CI | p-value |
|---|
| Histological subtype | | | | | | |
| NST vs. other | 0.82 | 0.28–2.44 | 0.7235 | 0.48 | 0.20–1.12 | 0.1088 |
| ILC vs. other | 0.46 | 0.13–1.68 | 0.238 | 0.25 | 0.07–0.85 | 0.0262 |
| Intrinsic molecular subtype | | | | | | |
| Luminal B HER2− vs. Luminal A | 2.35 | 0.91–6.05 | 0.078 | 0.76 | 0.22–2.59 | 0.662 |
| Luminal B HER2+ vs. Luminal A | 1.37 | 0.49–3.85 | 0.5495 | 1.02 | 0.29–3.59 | 0.9733 |
| HER2-type vs. Luminal A | 0.87 | 0.09–8.79 | 0.9034 | 0.89 | 0.13–6.33 | 0.9079 |
| TNBC vs. Luminal A | 1.41 | 0.41–4.84 | 0.5843 | 0.63 | 0.15–2.69 | 0.5279 |
| Ki-67 (high vs. low) | 0.3 | 0.12–0.73 | 0.0078 | 0.89 | 0.28–2.81 | 0.837 |
| Grade | | | | | | |
| 2 vs. 1 | 2.13 | 0.84–5.37 | 0.1106 | 1.85 | 0.71–4.80 | 0.2051 |
| 3 vs. 1 | 2.94 | 1.09–7.89 | 0.0326 | 3 | 1.09–8.24 | 0.0333 |
| PTEN (low vs. retained) | 0.13 | 0.07–0.24 | <0.0001 | 0.59 | 0.36–0.99 | 0.0435 |
Table 6.
Clinicopathologic features of the patients included in this study according to their biomarker status. All cases were re-classified, re-graded, and re-assessed for hormone receptor, Ki67, and HER2 status according to the latest guidelines. NST, no special type; TNBC, triple-negative breast cancer; *ER+/PR+/Ki67 low; #ER+/Ki67 high or ER+/PR-; ∞ER-/PR-/HER2+; §ER-/PR-/HER2-.
Table 6.
Clinicopathologic features of the patients included in this study according to their biomarker status. All cases were re-classified, re-graded, and re-assessed for hormone receptor, Ki67, and HER2 status according to the latest guidelines. NST, no special type; TNBC, triple-negative breast cancer; *ER+/PR+/Ki67 low; #ER+/Ki67 high or ER+/PR-; ∞ER-/PR-/HER2+; §ER-/PR-/HER2-.
| | ER+/HER2- | HER2+ | ER-/HER2- | Total |
|---|
| All patients, n (%) | 452 (74.3) | 97 (16.0) | 59 (9.7) | 608 (100) |
| Age at diagnosis, n (%) | | | | |
| ≥55 years | 322 (76.1) | 72 (17.0) | 29 (6.9) | 423 (69.6) |
| <55 years | 130 (70.3) | 25 (13.5) | 30 (16.2) | 185 (30.4) |
| Menopause, n (%) | | | | |
| Yes | 354 (75.0) | 82 (17.4) | 36 (7.6) | 472 (77.6) |
| No | 98 (72.1) | 15 (11.0) | 23 (16.9) | 136 (22.4) |
| Histological subtype, n (%) | | | | |
| Invasive carcinoma, NST | 354 (72.1) | 90 (18.3) | 47 (9.6) | 491 (80.7) |
| Lobular | 74 (93.7) | 5 (6.3) | 0 | 79 (13.0) |
| Other | 24 (63.2) | 2 (5.3) | 12 (31.5) | 38 (6.3) |
| Histological grade, n (%) | | | | |
| 1 | 65 (89.0) | 6 (8.2) | 2 (2.7) | 73 (12.0) |
| 2 | 233 (88.3) | 27 (10.2) | 4 (1.5) | 264 (43.4) |
| 3 | 154 (56.8) | 64 (23.6) | 53 (19.6) | 271 (44.6) |
| ER status, n (%) | | | | |
| Positive | 452 (84.0) | 86 (16.0) | 0 | 538 (88.4) |
| Negative | 0 | 11 (15.7) | 59 (84.3) | 70 (11.6) |
| PR status, n (%) | | | | |
| Positive | 398 (85.2) | 67 (14.3) | 2 (0.4) | 467 (76.8) |
| Negative | 54 (38.3) | 30 (21.2) | 57 (40.4) | 141 (23.2) |
| HER2 status, n (%) | | | | |
| Positive | 0 | 97 (100) | 0 | 97 (16.0) |
| Negative | 452 (88.5) | 0 | 59 (11.5) | 511 (84.0) |
| Ki67 status, n (%) | | | | |
| High | 262 (65.2) | 83 (20.6) | 57 (14.2) | 402 (66.1) |
| Low | 190 (92.2) | 14 (6.8) | 2 (1.0) | 206 (33.9) |
| Stage, n (%) | | | | |
| I | 212 (77.9) | 40 (14.7) | 20 (7.4) | 272 (44.7) |
| II | 164 (74.9) | 28 (12.8) | 27 (12.3) | 219 (36.0) |
| III-IV | 76 (65.0) | 29 (24.7) | 12 (10.3) | 117 (19.2) |
| Intrinsic molecular subtypes, n (%) | | | | |
| Luminal A* | 167 (100) | 0 | 0 | 167 (27.4) |
| Luminal B# | 285 (76.8) | 86 (23.2) | 0 | 371 (61.0) |
| HER2-type∞ | 0 | 11 (100) | 0 | 11 (1.8) |
| TNBC§ | 0 | 0 | 59 (100) | 59 (9.7) |