Reconstruction of the Evolutionary Histories of UGT Gene Superfamily in Legumes Clarifies the Functional Divergence of Duplicates in Specialized Metabolism
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of UGT Gene Family in Five Papilionoid Legumes
2.2. Phylogenetic Relationship of the UGTs in Five Papilionoid Legumes
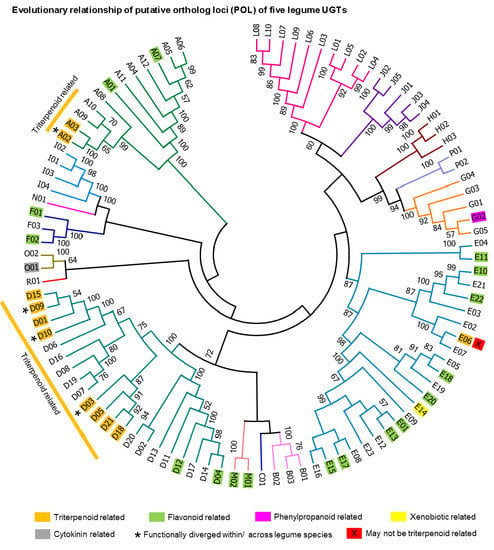
2.3. Putative Ortholog Loci Assignments for UGTs of Papilionoid Legumes
2.4. Expansion of UGTs in M. truncatula and G. max
2.5. Analysis of Intron Gain/Loss Events in M. truncatula and G. max
2.6. Chromosomal Locations and Gene Duplication Analyses in M. truncatula and G. max
2.7. Duplication History and Functional Divergence of Triterpene Related UGT POLs in M. truncatula and G. max
3. Discussion
3.1. Expansionary and Evolutionary Dynamics of the UGT Gene Family in M. truncatula and G. max: Insights from POL Assignments
3.2. Evolutionary Insights into the Sugar Chain Biosynthesis of Soyasaponins
3.2.1. Evolution of the C-3 Sugar Chain of Soyasaponins
3.2.2. Evolution of the C-22 Sugar Chain of Soyasaponins
3.3. Triterpene Related UGT POLs and Their Functional Divergence
4. Materials and Methods
4.1. Identification of Putative UGTs in Five Legumes
4.2. Phylogenetic Analysis
4.3. Assignment of POL for Legume UGTs
4.4. Estimation of Intron Addition or Deletion Events
4.5. Chromosomal Mapping, Gene Duplication, and Divergence Time Analyses
4.6. Microsynteny Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Schuman, B.; Alfaro, J.A.; Evans, S.V. Glycosyltransferase structure and function. In Topics in Current Chemistry; Peters, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 272, pp. 211–257. [Google Scholar]
- Wagner, G.K.; Pesnot, T. Glycosyltransferases and their assays. ChemBioChem 2010, 11, 1939–1949. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, D.M.; Liu, J.H.; Wu, H.; Wang, B.B.; Zhu, H.J.; Qiao, J.J. Glycosyltransferases: Mechanisms and applications in natural product development. Chem. Soc. Rev. 2015, 44, 8350–8374. [Google Scholar] [CrossRef] [PubMed]
- Osmani, S.A.; Bak, S.; Møller, B.L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 2009, 70, 325–347. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Gachon, C.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 2005, 10, 542–549. [Google Scholar] [CrossRef]
- Wang, X. Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 2009, 583, 3303–3309. [Google Scholar] [CrossRef] [Green Version]
- Bowles, D.; Isayenkova, J.; Lim, E.K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Härtl, K.; MacGraphery, K.; Rüdiger, J.; Schwab, W. Tailoring natural products with glycosyltransferases. In Biotechnology of Natural Products, 1st ed.; Schwab, W., Lange, B.M., Wüst, M., Eds.; Springer International Publishing AG: Heidelberg, Germany, 2018; pp. 219–263. [Google Scholar]
- Brazier-Hicks, M.; Offen, W.A.; Gershater, M.C.; Revett, T.J.; Lim, E.K.; Bowles, D.J.; Davies, G.J.; Edwards, R. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 20238–20243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Michlmayr, H.; Schweiger, W.; Malachova, A.; Shin, S.; Huang, Y.; Dong, Y.; Wiesenberger, G.; McCormick, S.; Lemmens, M.; et al. A barley UDP-glucosyltransferase inactivates nivalenol and provides Fusarium Head Blight resistance in transgenic wheat. J. Exp. Bot. 2017, 68, 2187–2197. [Google Scholar] [CrossRef] [PubMed]
- Goossens, A.; Osbourn, A.; Michoux, F.; Bak, S. Triterpene messages from the EU-FP7 project TriForC. Trends Plant Sci. 2018, 23, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A.; Goss, R.J.M.; Field, R.A. The saponins—Polar isoprenoids with important and diverse activities. Nat. Prod. Rep. 2011, 28, 1261–1268. [Google Scholar] [CrossRef]
- Seki, H.; Tamura, K.; Muranaka, T. Plant-derived isoprenoid sweetners: Recent progress in biosynthetic gene discovery and perspectives on microbial production. Biosci. Biotech. Biochem. 2018, 82, 927–934. [Google Scholar] [CrossRef] [Green Version]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O’Maille, P.; Osbourn, A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef]
- Salmon, M.; Thimmappa, R.B.; Minto, R.E.; Melton, R.E.; Hughes, R.K.; O’Maille, P.E.; Hemmings, A.M.; Osbourn, A. A conserved amino acid residue critical for product and substrate specificity in plant triterpene synthases. Proc. Natl. Acad. Sci. USA 2016, 113, E4407–E4414. [Google Scholar] [CrossRef] [Green Version]
- Xue, Z.; Duan, L.; Liu, D.; Guo, J.; Ge, S.; Dicks, J.; O’Maille, P.; Osbourn, A.; Qi, X. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 2012, 193, 1022–1038. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Fujisawa, Y.; Takahashi, Y.; Abe, H.; Yamane, K.; Mukaiyama, K.; Son, H.R.; Hiraga, S.; Kaga, A.; Anai, T.; et al. High throughput screening and characterization of a high-density soybean mutant library elucidate the biosynthesis pathway of triterpenoid saponins. Plant Cell Physiol. 2019, 60, 1082–1097. [Google Scholar] [CrossRef]
- Louveau, T.; Orme, A.; Pfalzgraf, H.; Stephenson, M.J.; Melton, R.; Saalbach, G.; Hemmings, A.M.; Leveau, A.; Rejzek, M.; Vickerstaff, R.J.; et al. Analysis of two new arabinosyltransferases belonging to the carbohydrate-active enzyme (CAZY) glycosyl transferase family 1 provides insights into disease resistance and sugar donor specificity. Plant Cell 2018, 30, 3038–3057. [Google Scholar] [CrossRef] [Green Version]
- Augustin, J.M.; Drok, S.; Shinoda, T.; Sanmiya, K.; Nielsen, J.K.; Khakimov, B.; Olsen, C.E.; Hansen, E.H.; Kuzina, V.; Ekstrøm, C.T.; et al. UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyse sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol. 2012, 160, 1881–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guang, C.; Chen, J.; Sang, S.; Cheng, S. Biological functionality of soyasaponins and soyasapogenols. J. Agri. Food Chem. 2014, 62, 8247–8255. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Tsurumi, S. The unique auxin influx modulator chromosaponin I: A physiological overview. Plant Tissue Cult. 2002, 12, 181–194. [Google Scholar]
- Yano, R.; Takagi, K.; Takada, Y.; Mukaiyama, K.; Tsukamoto, C.; Sayama, T.; Kaga, A.; Anai, T.; Sawai, S.; Ohyama, K.; et al. Metabolic switching of astringent and beneficial triterpenoid saponins in soybean is achieved by a loss-of-function mutation in cytochrome P450 72A69. Plant, J. 2017, 89, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M.; Nishimura, K.; Yasuyama, N.; Ebizuka, Y. Identification and characterization of glycosyltransferases involved in the biosynthesis of soyasaponin I in Glycine max. FEBS Lett. 2010, 584, 2258–2264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayama, T.; Ono, E.; Takagi, K.; Takada, Y.; Horikawa, M.; Nakamoto, Y.; Hirose, A.; Sasama, H.; Ohashi, M.; Hasegawa, H.; et al. The Sg-1 glycosyltransferase locus regulates structural diversity of triterpenoid saponins of soybean. Plant Cell 2012, 24, 2123–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, K.; Yano, R.; Tochigi, S.; Fujisawa, Y.; Tsuchinaga, H.; Takahashi, Y.; Takada, Y.; Kaga, A.; Anai, T.; Tsukamoto, C.; et al. Genetic and functional characterization of Sg-4 glycosyltransferase involved in the formation of sugar chain structure at the C-3 position of soybean saponins. Phytochemistry 2018, 156, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Yano, R.; Takagi, K.; Tochigi, S.; Fujisawa, Y.; Nomura, Y.; Tsuchinaga, H.; Takahashi, Y.; Takada, Y.; Kaga, A.; Anai, T.; et al. Isolation and characterization of the soybean Sg-3 gene that is involved in genetic variation in sugar chain composition at the C-3 position in soyasaponins. Plant Cell Physiol. 2018, 59, 792–805. [Google Scholar] [CrossRef] [Green Version]
- Sundaramoorthy, J.; Par, G.T.; Komagamine, K.; Tsukamoto, C.; Chang, J.H.; Lee, J.D.; Kim, J.H.; Seo, H.S.; Song, J.T. Biosynthesis of DDMP saponins in soybean is regulated by a distinct UDP-glycosyltransferase. New Phytol. 2019, 222, 261–274. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plant species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef] [Green Version]
- Wink, M. Evolution of secondary metabolites in legumes (Fabaceae). S. Afr. J. Bot. 2013, 89, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, P.; Li, Y.; Liu, Y.; Yu, J.; Ma, X.; Sun, S.; Yang, N.; Xia, R.; Lei, T.; et al. Hierarchically aligning 10 legume genomes establishes family-level genomics platform. Plant Physiol. 2017, 174, 284–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Yao, S.; Dai, X.; Yin, Q.; Liu, Y.; Jiang, X.; Wu, Y.; Qian, Y.; Pang, Q.; Gao, L.; et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 2016, 67, 2285–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genomics 2012, 13, 175. [Google Scholar] [CrossRef] [Green Version]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Muller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Niu, L.; Yang, Q.; Dong, B.; Wang, L.; Dong, M.; Fan, X.; Jian, Y.; Meng, D.; Fu, Y. Genome-wide identification and characterization of UGT family in pigeonpea (Cajanus cajan) and expression analysis in abiotic stress. Trees 2019, 33, 987–1002. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Bao, L.; Shah, Z.H.; Lee, J.M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhuo, X.; Yan, Z.; Zhang, Q. Comparative genomic and transcriptomic analyses of family-1 UDP glycosyltransferase in Prunus mume. Int. J. Mol. Sci. 2018, 19, 3382. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Shen, G.; Di, S.; Fan, C.; Chang, Z.; Pang, Y. Genome-wide identification and functional characterization of UDP-glucosyltransferase genes involved in flavonoid biosynthesis in Glycine max. Plant Cell Physiol. 2017, 58, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Shen, G.; Chang, Z.; Tang, Y.; Gao, H.; Pang, Y. Involvement of three putative glucosyltransferases from the UGT72 family in flavonol glucoside/rhamnoside biosynthesis in Lotus japonicus seeds. J. Exp. Bot. 2017, 68, 597–612. [Google Scholar] [PubMed] [Green Version]
- Tomcal, M.; Stiffler, N.; Barkan, A. POGs2: A web portal to facilitate cross-species inferences about protein architecture and function in plants. PLoS ONE 2013, 8, e82569. [Google Scholar] [CrossRef]
- Van Bel, M.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van de Peer, Y.; Coppens, F.; Vandepoele, K. PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2017, 46, D1190–D1196. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Dash, S.; Campbell, J.D.; Cannon, E.K.; Cleary, A.M.; Huang, W.; Kalberer, S.R.; Karingula, V.; Rice, A.G.; Singh, J.; Umale, P.E.; et al. Legume information system (LegumeInfo.org): A key component of a set of federated data resources for the legume family. Nucleic Acids Res. 2016, 44, D1181–D1188. [Google Scholar] [CrossRef] [Green Version]
- Naoumkina, M.A.; Modolo, L.V.; Huhman, D.V.; Urbanczyk-Wochniak, E.; Tang, Y.; Sumner, L.W.; Dixon, R.A. Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 2010, 22, 850–866. [Google Scholar] [CrossRef] [Green Version]
- Ishimoto M’s Research Group. Identification of UGTs involved in soyasaponin glycosylation. Unpublished Work.
- Achnine, L.; Huhman, D.V.; Farag, M.A.; Sumner, L.W.; Blount, J.W.; Dixon, R.A. Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 2005, 41, 875–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proost, S.; Van Bel, M.; Sterck, L.; Billiau, K.; Van Parys, T.; Van de Peer, Y.; Vandepoele, K. PLAZA: A comparative genomics resource to study gene and genome evolution in plants. Plant Cell 2009, 21, 3718–3731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trachana, K.; Larsson, T.A.; Powell, S.; Chen, W.H.; Doerks, T.; Muller, J.; Bork, P. Orthology prediction methods: A quality assessment using curated protein families. Bioessays 2011, 33, 769–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, N.S.; Stiffler, N.; Barkan, A. POGs/PlantRBPs: A resource for comparative genomics in plants. Nucl. Acid Res. 2007, 55, D852–D856. [Google Scholar] [CrossRef]
- De Vega, J.J.; Ayling, S.; Hegarty, M.; Kudrna, D.; Goicoechea, J.L.; Ergon, A.; Rognli, O.A.; Jones, C.; Swain, M.; Geurts, R.; et al. Red clover (Trifolium pretense L.) draft genome provides a platform for trait improvement. Sci. Rep. 2015, 5, 17394. [Google Scholar] [CrossRef]
- Ober, D. Seeing double: Gene duplication and diversification in plant secondary metabolism. Trends. Plant Sci. 2005, 10, 444–449. [Google Scholar] [CrossRef]
- Hamberger, B.; Bak, S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. B 2013, 368, 20120426. [Google Scholar] [CrossRef] [Green Version]
- Okubo, K.; Yoshiki, Y. Oxygen-radical-scavenging activity of DDMP-conjugated saponins and physiological role in leguminous plant. In Saponins Used in Food and Agriculture; Waller, G.R., Yamasaki, K., Eds.; Plenum Press: New York, NY, USA, 1996; Volume 405, pp. 141–154. [Google Scholar]
- Pollier, J.; Morreel, K.; Geelen, D.; Goossens, A. Metabolite profiling of triterpene saponins in Medicago truncatula hairy roots by liquid chromatography fourier transform ion cyclotron resonance mass spectrometry. J. Nat. Prod. 2011, 74, 1462–1476. [Google Scholar] [CrossRef] [Green Version]
- Kinjo, J.; Kishida, F.; Watanabe, K.; Hashimoto, F.; Nohara, T. Five new triterpene glycosides from Russell lupine. Chem. Parm. Bull. 1994, 42, 1874–1878. [Google Scholar] [CrossRef] [Green Version]
- Dhaubhadel, S.; Farhangkhoee, M.; Chapman, R. Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J. Exp. Bot. 2004, 59, 981–994. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Cai, W.; Gao, W.; Liu, C. A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. New Phytol. 2016, 212, 123–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Chen, K.; Hu, Z.M.; Li, K.; Song, W.; Yu, L.Y.; Leung, C.H.; Ma, D.L.; Qiao, X.; Ye, M. UGT73F17, a new glycosyltransferase from Glycyrrhiza uralensis, catalyzes the regiospecific glycosylation of pentacyclic triterpenoids. Chem. Commun. 2018, 54, 8594–8597. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Kato, T.; Nakao, M.; Sasamoto, S.; Watanabe, A.; Ono, A.; Kawashima, K.; et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008, 15, 227–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wemersson, R.; Pedersen, A.G. RevTrans-Constructing alignments of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003, 31, 3537–3539. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Young, N.D.; Debelle, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, M.; Misra, G.; Patel, R.K.; Priya, P.; Jhanwar, S.; Khan, A.W.; Shah, N.; Singh, V.K.; Garg, R.; Jeena, G.; et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 2013, 74, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Cleary, A.; Farmer, A. Genome Context Viewer: Visual exploration of multiple annotated genomes using microsynteny. Bioinformatics 2018, 34, 1562–1564. [Google Scholar] [CrossRef] [Green Version]






| No. | Plant Species Name * | No. UGTs in Different Phylogenetic Groups | Total UGTs | Ref. | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | ||||
| 1 | Mimulus guttatus | 10 | 2 | 3 | 11 | 14 | – | 12 | 1 | 4 | 2 | 9 | 17 | 2 | 1 | 9 | 3 | – | – | 100 | [36] |
| 2 | Camellia sinensis | 15 | 5 | 2 | 20 | 23 | 2 | 13 | 2 | 2 | 2 | 1 | 27 | 3 | – | 6 | 6 | – | 3 | 132 | [37] |
| 3 | Vitis vinifera | 23 | 3 | 4 | 8 | 46 | 5 | 15 | 7 | 14 | 4 | 2 | 31 | 5 | 1 | 2 | 11 | – | – | 181 | [36] |
| 4 | Linum usitatissimum | 16 | 5 | 6 | 21 | 22 | 1 | 19 | 6 | 9 | 4 | 5 | 19 | 3 | 1 | – | – | – | – | 137 | [38] |
| 5 | Populus trichocarpa | 12 | 2 | 6 | 14 | 49 | – | 42 | 5 | 5 | 6 | 2 | 23 | 6 | 1 | 3 | 2 | – | – | 178 | [36] |
| 6 | Cucumis sativus | 10 | 1 | 2 | 12 | 13 | – | 11 | 5 | – | 2 | 1 | 17 | 2 | 1 | 3 | 5 | – | – | 85 | [36] |
| 7 | Arabidopsis thaliana | 14 | 3 | 3 | 13 | 22 | 3 | 6 | 19 | 1 | 2 | 2 | 17 | 1 | 1 | – | – | – | – | 107 | [36] |
| 8 | Brassica rapa | 12 | 4 | 4 | 24 | 31 | 1 | 9 | 18 | 1 | 3 | 3 | 26 | 2 | 2 | – | – | – | – | 140 | [39] |
| 9 | Brassica napus | 17 | 10 | 10 | 36 | 48 | 2 | 14 | 35 | 2 | 3 | 6 | 61 | 4 | 3 | – | – | – | – | 251 | [39] |
| 10 | Brassica oleraca | 15 | 7 | 4 | 23 | 32 | – | 8 | 23 | 1 | 2 | 3 | 32 | 2 | 2 | – | – | – | – | 154 | [39] |
| 11 | Cajanus Cajan | 2 | 2 | 1 | 36 | 33 | – | 9 | – | 5 | – | – | 12 | 2 | – | 6 | 12 | – | – | 120 | [40] |
| 12 | Glycine max | 25 | 3 | 1 | 43 | 36 | 1 | 15 | 3 | 18 | 3 | 2 | 19 | 4 | 1 | 5 | 3 | – | – | 182 | [36] |
| Glycine max | 5 | 1 | 2 | 38 | 46 | 6 | 16 | 2 | 4 | – | – | 18 | 5 | – | 6 | – | – | – | 149 | [41] | |
| Glycine max | 21 | 3 | – | 46 | 52 | 8 | 16 | 3 | 17 | 7 | – | 19 | 5 | 1 | 6 | 4 | – | – | 208 | This study | |
| 13 | Phaseolus vulgaris | 19 | 3 | 2 | 33 | 33 | 5 | 18 | 3 | 15 | 3 | – | 17 | 4 | 1 | 6 | 5 | – | 1 | 168 | This study |
| 14 | Lotus japonicus | 9 | 3 | – | 25 | 22 | 2 | 9 | 1 | 2 | 1 | 0 | 10 | 1 | 1 | 6 | 1 | – | 1 | 94 | This study |
| 15 | Medicago truncatula | 28 | 4 | – | 55 | 55 | 2 | 39 | 3 | 5 | 9 | – | 33 | 2 | 1 | 3 | 3 | – | 1 | 243 | This study |
| 16 | Trifolium pratense | 11 | 3 | – | 29 | 39 | 1 | 13 | 3 | 1 | 2 | – | 12 | 1 | – | 2 | 3 | – | 1 | 121 | This study |
| 17 | Malus domestica | 33 | 4 | 7 | 13 | 55 | 6 | 40 | 14 | 11 | 12 | 6 | 16 | 13 | 1 | 5 | 5 | – | – | 241 | [36] |
| 18 | Prunus mume | 16 | 2 | 3 | 17 | 23 | 3 | 18 | 10 | 4 | ? | 8 | 17 | 3 | ? | – | – | – | – | 130 | [42] |
| 19 | Prunus persica | 10 | 2 | 4 | 19 | 29 | 4 | 34 | 9 | 5 | 7 | 7 | 18 | 14 | 1 | 1 | 4 | – | – | 168 | [43] |
| 20 | Oryza sativa | 14 | 9 | 8 | 26 | 38 | – | 20 | 7 | 9 | 3 | 1 | 23 | 5 | 2 | 6 | 9 | – | – | 180 | [36] |
| 21 | Triticum aestivum | 22 | 3 | 2 | 17 | 37 | 2 | 4 | 5 | 7 | 5 | – | 19 | 3 | 1 | 3 | 13 | 36 | – | 179 | [44] |
| 22 | Sorghum bicolor | 10 | 4 | 6 | 24 | 50 | – | 17 | 12 | 8 | 3 | 1 | 26 | 6 | 3 | 8 | 2 | – | – | 180 | [36] |
| 23 | Zea mays | 8 | 3 | 5 | 18 | 34 | 2 | 12 | 9 | 9 | 3 | 1 | 23 | 3 | 4 | 5 | 1 | 7 | – | 147 | [45] |
| Phylogenetic Groups | Distribution of UGT POLs among the Phylogenetic Groups a | |||||
|---|---|---|---|---|---|---|
| M. truncatula | G. max | P. vulgaris | L. japonicus | T. pratense | Total | |
| A | 10 (28) | 10 (21) | 09 (19) | 08 (09) | 07 (11) | 12 |
| B | 03 (04) | 03 (03) | 02 (03) | 02 (03) | 03 (03) | 03 |
| C | – | – | 01 (02) | – | – | 01 |
| D | 14 (55) | 19 (46) | 16 (33) | 15 (25) | 13 (29) | 21 |
| E | 18 (55) | 21 (52) | 16 (33) | 12 (22) | 14 (39) | 23 |
| F | 01 (02) | 03 (08) | 03 (05) | 02 (02) | 01 (01) | 03 |
| G | 05 (39) | 04 (16) | 03 (18) | 03 (09) | 04 (13) | 05 |
| H | 03 (03) | 02 (03) | 03 (03) | 01 (01) | 03 (03) | 03 |
| I | 02 (05) | 04 (17) | 04 (15) | 02 (02) | 01 (01) | 04 |
| J | 04 (09) | 05 (07) | 02 (03) | 01 (01) | 02 (02) | 05 |
| K | – | – | – | – | – | – |
| L | 09 (33) | 08 (19) | 09 (17) | 07 (10) | 07 (12) | 10 |
| M | 02 (02) | 02 (05) | 02 (04) | 01 (01) | 01 (01) | 02 |
| N | 01 (01) | 01 (01) | 01 (01) | 01 (01) | - | 01 |
| O | 02 (03) | 02 (06) | 02 (06) | 02 (06) | 02 (02) | 02 |
| P | 01 (03) | 02 (04) | 02 (05) | 01 (01) | 01 (03) | 02 |
| Q | – | – | – | – | – | – |
| R | 01 (01) | – | 01 (01) | 01 (01) | 01 (01) | 01 |
| Total | 76 (243) | 86 (208) | 76 (168) | 59 (94) | 60 (121) | 98 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnamurthy, P.; Tsukamoto, C.; Ishimoto, M. Reconstruction of the Evolutionary Histories of UGT Gene Superfamily in Legumes Clarifies the Functional Divergence of Duplicates in Specialized Metabolism. Int. J. Mol. Sci. 2020, 21, 1855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051855
Krishnamurthy P, Tsukamoto C, Ishimoto M. Reconstruction of the Evolutionary Histories of UGT Gene Superfamily in Legumes Clarifies the Functional Divergence of Duplicates in Specialized Metabolism. International Journal of Molecular Sciences. 2020; 21(5):1855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051855
Chicago/Turabian StyleKrishnamurthy, Panneerselvam, Chigen Tsukamoto, and Masao Ishimoto. 2020. "Reconstruction of the Evolutionary Histories of UGT Gene Superfamily in Legumes Clarifies the Functional Divergence of Duplicates in Specialized Metabolism" International Journal of Molecular Sciences 21, no. 5: 1855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051855