Role of Chitin Deacetylase 1 in the Molting and Metamorphosis of the Cigarette Beetle Lasioderma serricorne
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of LsCDA1
2.2. Developmental and Tissue-Specific Expression of LsCDA1
2.3. Expression of LsCDA1 in Response to 20E Signaling
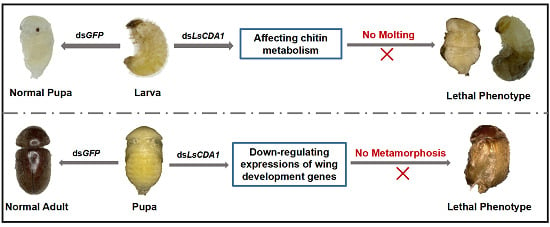
2.4. Knockdown of LsCDA1 Impairs Larval–Pupal and Pupal–Adult Transitions
3. Discussion
3.1. Characteristics and Expression Profiles of LsCDA1
3.2. Transcriptional Regulation of LsCDA1 by 20E
3.3. Knockdown of LsCDA1 Affects Chitin Metabolism and Impairs Molting
3.4. The Function of LsCDA1 Involved in Wing Development During Pupal–Adult Metamorphosis
4. Materials and Methods
4.1. Insects
4.2. Molecular Cloning and Bioinformatic Analysis
4.3. Developmental and Tissue-Specific Expression Analysis of LsCDA1
4.4. Expression of LsCDA1 after 20E Treatment and RNAi of Ecdysone Synthesis and Signaling Genes
4.5. Function Analysis of LsCDA1 by RNAi
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Choi, H.K.; Choi, K.H.; Kramer, K.J.; Muthukrishnan, S. Isolation and characterization of a genomic clone for the gene of an insect molting enzyme, chitinase. Insect Biochem. Mol. Biol. 1997, 27, 37–47. [Google Scholar] [CrossRef]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moussian, B.; Schwarz, H.; Bartoszewski, S.; Nusslein-Volhard, C. Involvement of chitin in exoskeleton morphogenesis in Drosophila melanogaster. J. Morphol. 2005, 264, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Nation, J.L. Insect Physiology and Biochemistry, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Cohen, E. Chitin synthesis and inhibition: A revisit. Pest Manag. Sci. 2001, 57, 946–950. [Google Scholar] [CrossRef]
- Kramer, K.; Muthukrishnan, S. Chitin metabolism in insects. Compr. Mol. Insect Sci. 2005, 4, 111–144. [Google Scholar]
- Tetreau, G.; Cao, X.L.; Chen, Y.R.; Muthukrishnan, S.; Jiang, H.B.; Blissard, G.W.; Kanost, M.R.; Wang, P. Overview of chitin metabolism enzymes in Manduca sexta: Identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. Biol. 2015, 62, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Xi, Y.; Pan, P.L.; Ye, Y.X.; Yu, B.; Xu, H.J.; Zhang, C.X. Chitinase-like gene family in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2015, 24, 29–40. [Google Scholar] [CrossRef]
- Xi, Y.; Pan, P.L.; Zhang, C.X. The β-N-acetylhexosaminidase gene family in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2015, 24, 601–610. [Google Scholar] [CrossRef]
- Xi, Y.; Pan, P.L.; Ye, Y.X.; Yu, B.; Zhang, C.X. Chitin deacetylase family genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 2014, 23, 695–705. [Google Scholar] [CrossRef]
- Tsigos, I.; Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 2000, 18, 305–312. [Google Scholar] [CrossRef]
- Guo, W.; Li, G.; Pang, Y.; Wang, P. A novel chitin binding protein identified from the peritrophic membrane of the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 2005, 35, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Dixit, R.; Begum, K.; Park, Y.; Specht, C.A.; Merzendorfer, H.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 2009, 39, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Xu, K.K.; Yan, X.; Chen, C.X.; Cao, Y.; Meng, Y.L.; Li, C. Functional characterization of chitin deacetylase 1 gene disrupting larval–pupal transition in the drugstore beetle using RNA interference. Comp. Biochem. Physiol. B 2018, 219–220, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Mu, L.L.; Chen, Z.C.; Fu, K.Y.; Guo, W.C.; Li, C.; Li, G.Q. Disruption of ecdysis in Leptinotarsa decemlineata by knockdown of chitin deacetylase 1. J. Asia Pac. Entomol. 2019, 22, 443–452. [Google Scholar] [CrossRef]
- Wu, J.J.; Chen, Z.C.; Wang, Y.W.; Fu, K.Y.; Guo, W.C.; Li, G.Q. Silencing chitin deacetylase 2 impairs larval–pupal and pupal-adult molts in Leptinotarsa decemlineata. Insect Mol. Biol. 2019, 28, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008, 38, 440–451. [Google Scholar] [CrossRef] [Green Version]
- Ding, G.W.; Yu, R.R.; Yang, M.L.; Ma, E.B.; Yang, J.; Zhang, J.Z. Molecular characterization and functional analysis of chitin deacetylase 1 gene in Oxya chinensis (Orthoptera: Acrididae). Acta Entomol. Sin. 2014, 57, 1265–1271. [Google Scholar]
- Yu, R.R.; Liu, W.M.; Zhao, X.M.; Zhang, M.; Li, D.Q.; Zuber, R.; Ma, E.B.; Zhu, K.Y.; Moussian, B.; Zhang, J.Z. LmCDA1 organizes the cuticle by chitin deacetylation in Locusta migratoria. Insect Mol. Biol. 2019, 28, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Luschnig, S.; Batz, T.; Armbruster, K.; Krasnow, M.A. Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 2006, 16, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.H.; Li, H.F.; Yang, Y.; Yang, R.L.; Yang, W.J.; Jiang, H.B.; Dou, W.; Smagghe, G.; Wang, J.J. Genome-wide identification of chitinase and chitin deacetylase gene families in the oriental fruit fly, Bactrocera dorsalis (Hendel). Comp. Biochem. Physiol. D 2018, 27, 13–22. [Google Scholar] [CrossRef]
- Quan, G.; Ladd, T.; Duan, J.; Wen, F.; Doucet, D.; Cusson, M.; Krell, P.J. Characterization of a spruce budworm chitin deacetylase gene: Stage- and tissue-specific expression, and inhibition using RNA interference. Insect Biochem. Mol. Biol. 2013, 43, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Li, X.; Zhang, T.; Zhu, X.; Li, J. Cloning and tissue-specific expression of a chitin deacetylase gene from Helicoverpa armigera (Lepidoptera: Noctuidae) and its response to Bacillus thuringiensis. J. Insect Sci. 2015, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.P.; Zhao, D.; Zhang, Y.K.; Guo, W.; Wang, W.; Zhao, K.L.; Gao, Y.J.; Wang, X.Y. Identification and characterization of chitin deacetylase 2 from the American white moth, Hyphantria cunea (Drury). Gene 2018, 670, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Yan, J.M.; Liu, Q.; Zhang, Y.H.; Gong, J.; Hou, Y. Genome-wide analysis and hormone regulation of chitin deacetylases in silkworm. Int. J. Mol. Sci. 2019, 20, 1679. [Google Scholar] [CrossRef] [Green Version]
- Sandoval-Mojica, A.F.; Scharf, M.E. Gut genes associated with the peritrophic matrix in Reticulitermes flavipes (Blattodea: Rhinotermitidae): Identification and characterization. Arch. Insect Biochem. Physiol. 2016, 92, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Cheng, J.; Lyu, Z.H.; Li, Z.X.; Chen, J.X.; Lin, T. Chitin deacetylase 1 and 2 are indispensable for larval–pupal and pupal–adult molts in Heortia vitessoides (Lepidoptera: Crambidae). Comp. Biochem. Physiol. B 2019, 237, 110325. [Google Scholar] [CrossRef]
- Wang, S.; Jayaram, S.J.; Senti, K.; Tsarouhas, V.; Jin, H.; Samakovlis, C. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 2006, 16, 180–185. [Google Scholar] [CrossRef] [Green Version]
- Gangishetti, U.; Veerkamp, J.; Bezdan, D.; Schwarz, H.; Lohmann, I.; Moussian, B. The transcription factor grainy head and the steroid hormone ecdysone cooperate during differentiation of the skin of Drosophila melanogaster. Insect Mol. Biol. 2012, 21, 283–295. [Google Scholar] [CrossRef]
- Zhang, M.; Ji, Y.N.; Zhang, X.B.; Ma, P.J.; Wang, Y.W.; Moussian, B.; Zhang, J.Z. The putative chitin deacetylases Serpentine and Vermiform have non-redundant functions during Drosophila wing development. Insect Biochem. Mol. Biol. 2019, 110, 128–135. [Google Scholar] [CrossRef]
- Toprak, U.; Baldwin, D.; Erlandson, M.; Gillott, C.; Hou, X.; Coutu, C.; Hegedus, D.D. A chitin deacetylase and putative insect intestinal lipases are components of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix. Insect Mol. Biol. 2010, 17, 573–585. [Google Scholar] [CrossRef]
- Zhong, X.W.; Wang, X.H.; Tan, X.; Xia, Q.Y.; Xiang, Z.H.; Zhao, P. Identification and molecular characterization of a chitin deacetylase from Bombyx mori peritrophic membrane. Int. J. Mol. Sci. 2014, 15, 1946–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahroof, R.M.; Phillips, T.W. Life history parameters of Lasioderma serricorne (F.) as influenced by food sources. J. Stored Prod. Res. 2008, 44, 219–226. [Google Scholar] [CrossRef]
- Li, C.; Li, Z.Z.; Cao, Y.; Zhou, B.; Zheng, X.W. Partial characterization of stress-induced carboxylesterase from adults of Stegobium paniceum and Lasioderma serricorne (Coleoptera: Anobiidae) subjected to CO2-enriched atmosphere. J. Pest Sci. 2009, 82, 7–11. [Google Scholar] [CrossRef]
- Minor, M.F. Do adult cigarette beetle feed? Tob. Sci. 1979, 23, 61–64. [Google Scholar]
- Riudavets, J.; Salas, I.; Pons, M.J. Damage characteristics produced by insect pests in packaging film. J. Stored Prod. Res. 2007, 43, 564–570. [Google Scholar] [CrossRef]
- Collins, D.; Conyers, S. The effect of sub-zero temperatures on different lifestages of Lasioderma serricorne (F.) and Ephestia elutella (Hübner). J. Stored Prod. Res. 2010, 46, 234–241. [Google Scholar] [CrossRef]
- Yu, C.; Subramanyam, B.; Flinn, P.W.; Gwirtz, J.A. Susceptibility of Lasioderma serricorne (Coleoptera: Anobiidae) life stages to elevated temperatures used during structural heat treatments. J. Econ. Entomol. 2011, 104, 317–324. [Google Scholar] [CrossRef]
- Imai, T. The additive effect of carbon dioxide on mortality of the cigarette beetle Lasioderma serricorne (Coleoptera: Anobiidae) in low-oxygen atmospheres. Appl. Entomol. Zool. 2015, 50, 11–15. [Google Scholar] [CrossRef]
- Saeed, M.B.E.E.E.M.; Laing, M.D.; Miller, R.M.; Bancole, B. Ovicidal, larvicidal and insecticidal activity of strains of Beauveria bassiana (Balsamo) Vuillemin against the cigarette beetle, Lasioderma serricorne Fabricius (Coleoptera: Anobiidae), on rice grain. J. Stored Prod. Res. 2017, 74, 78–86. [Google Scholar] [CrossRef]
- Hironaka, M.; Kamura, T.; Osada, M.; Sasaki, R.; Shinoda, K.; Hariyama, T.; Miyatake, T. Adults of Lasioderma serricorne and Stegobium paniceum (Anobiidae: Coleoptera) are attracted to ultraviolet (UV) over blue light LEDs. J. Econ. Entomol. 2017, 110, 1911–1915. [Google Scholar] [CrossRef]
- Rajendran, S.; Narasimhan, K.S. Phosphine resistance in the cigarette beetle Lasioderma serricorne (Coleoptera: Anobiidae) and overcoming control failures during fumigation of stored tobacco. Int. J. Pest Manag. 1994, 40, 207–210. [Google Scholar] [CrossRef]
- Sağlam, Ö.; Edde, P.A.; Phillips, T.W. Resistance of Lasioderma serricorne (Coleoptera: Anobiidae) to fumigation with phosphine. J. Econ. Entomol. 2015, 108, 2489–2495. [Google Scholar] [CrossRef] [PubMed]
- Abdelghany, A.Y.; Awadalla, S.S.; Abdel-Baky, N.F.; EL-Syrafi, H.A.; Fields, P.G. Efficacy of reduced risk insecticides on penetration into jute and polyethylene bags by Lasioderma serricorne (F.) (Coleoptera: Anobiidae). J. Stored Prod. Res. 2016, 69, 190–194. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, W.J.; Huang, D.Y.; Wang, Y.; Wei, J.Y.; Li, Z.H.; Sun, J.S.; Bai, J.F.; Tian, Z.F.; Wang, P.J.; et al. Chemical compositions and insecticidal activities of Alpinia kwangsiensis essential oil against Lasioderma serricorne. Molecules 2015, 20, 21939–21945. [Google Scholar] [CrossRef]
- Cohen, E. Chitin synthesis and degradation as targets for pesticide action. Arch. Insect Biochem. Physiol. 1993, 22, 245–261. [Google Scholar] [CrossRef]
- Chen, X.Y.L.; Xu, K.K.; Yan, X.; Chen, C.X.; Cao, Y.; Wang, Y.W.; Li, C.; Yang, W.J. Characterization of a β-N-acetylglucosaminidase gene and its involvement in the development of Lasioderma serricorne (Fabricius). J. Stored Prod. Res. 2018, 77, 156–165. [Google Scholar] [CrossRef]
- Yang, W.J.; Xu, K.K.; Yan, Y.; Li, C. Knockdown of β-N-acetylglucosaminidase 2 impairs molting and wing development in Lasioderma serricorne (Fabricius). Insects 2019, 10, 396. [Google Scholar] [CrossRef] [Green Version]
- Blair, D.E.; Schuttelkopf, A.W.; MacRae, J.I.; vanAalten, D.M. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.Z.; Liu, M.H.; Wang, X.Y.; Yang, X.; Wang, W.L.; Geng, L.; Yu, D.; Liu, X.L.; Xu, J.P. Identification and expression profiles of chitin deacetylase genes in the rice leaf folder, Cnaphalocrocis medinalis. J. Asia Pac. Entomol. 2016, 19, 691–696. [Google Scholar] [CrossRef]
- Luan, J.B.; Ghanim, M.; Liu, S.S.; Czosnek, H. Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 2013, 43, 740–746. [Google Scholar] [CrossRef]
- Niwa, R.; Niwa, Y.S. Enzymes for ecdysteroid biosynthesis: Their biological functions in insects and beyond. Biosci. Biotechnol. Biochem. 2014, 78, 1283–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, R.J.; Billas, I.M.; Bonneton, F.; Graham, L.D.; Lawrence, M.C. Ecdysone receptors: From the Ashburner model to structural biology. Annu. Rev. Entomol. 2013, 58, 251–271. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Zhang, D.W.; Tang, B.; Chen, J.; Chen, J.; Lu, L.; Zhang, W.Q. Identification of 20-hydroxyecdysone late-response genes in the chitin biosynthesis pathway. PLoS ONE 2010, 5, e14058. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Q.; Zhang, J.Q.; Wang, Y.; Liu, X.J.; Ma, E.B.; Sun, Y.; Li, S.; Zhu, K.Y.; Zhang, J.Z. Two chitinase 5 genes from Locusta migratoria: Molecular characteristics and functional differentiation. Insect Biochem. Mol. Biol. 2015, 58, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Li, D.Q.; Zhang, X.Y.; Li, S.; Zhu, K.Y.; Guo, Y.P.; Ma, E.B.; Zhang, J.Z. RNA interference to reveal roles of β-N-acetylglucosaminidase gene during molting process in Locusta migratoria. Insect Sci. 2013, 20, 109–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.J.; Xu, K.K.; Cong, L.; Wang, J.J. Identification, mRNA expression, and functional analysis of chitin synthase 1 gene and its two alternative splicing variants in oriental fruit fly, Bactrocera dorsalis. Int. J. Biol. Sci. 2013, 9, 331–342. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, W.W.; Fang, Y.; Kong, L.F.; Li, X.; Sima, Y.H.; Xu, S.Q. Chitin synthase A: A novel epidermal development regulation gene in the larvae of Bombyx mori. Mol. Biol. Rep. 2014, 41, 4177–4186. [Google Scholar] [CrossRef]
- Shi, J.F.; Mu, L.L.; Guo, W.C.; Li, G.Q. Identification and hormone induction of putative chitin synthase genes and splice variants in Leptinotarsa decemlineata (SAY). Arch. Insect Biochem. Physiol. 2016, 92, 242–258. [Google Scholar] [CrossRef]
- Lyu, Z.H.; Chen, J.X.; Li, Z.X.; Cheng, J.; Wang, C.Y.; Lin, T. Knockdown of β-N-acetylglucosaminidase gene disrupts molting process in Heortia vitessoides Moore. Arch. Insect Biochem. Physiol. 2019, 101, e21561. [Google Scholar] [CrossRef]
- Chen, C.; Yang, H.; Tang, B.; Yang, W.J.; Jin, D.C. Identification and functional analysis of chitinase 7 gene in white-backed planthopper, Sogatella furcifera. Comp. Biochem. Physiol. B 2017, 208–209, 19–28. [Google Scholar] [CrossRef]
- Clark-Hachtel, C.M.; Linz, D.M.; Tomoyasu, Y. Insights into insect wing origin provided by functional analysis of vestigial in the red flour beetle, Tribolium castaneum. Proc. Natl. Acad. Sci. USA 2013, 110, 16951–16956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Li, B.; Ma, S.; Lü, P.; Chen, K. Dusky works upstream of Four-jointed and Forked in wing morphogenesis in Tribolium castaneum. Insect Mol. Biol. 2017, 26, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.J.; Xu, K.K.; Cao, Y.; Meng, Y.L.; Liu, Y.; Li, C. Identification and expression analysis of four small heat shock protein genes in cigarette beetle, Lasioderma serricorne (Fabricius). Insects 2019, 10, 139. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemas, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.J.; Chen, C.X.; Yan, Y.; Xu, K.K.; Li, C. Clip-domain serine protease gene (LsCLIP3) is essential for larval–pupal molting and immunity in Lasioderma serricorne. Front. Physiol. 2020, 10, 1631. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.-J.; Xu, K.-K.; Yan, Y.; Li, C.; Jin, D.-C. Role of Chitin Deacetylase 1 in the Molting and Metamorphosis of the Cigarette Beetle Lasioderma serricorne. Int. J. Mol. Sci. 2020, 21, 2449. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072449
Yang W-J, Xu K-K, Yan Y, Li C, Jin D-C. Role of Chitin Deacetylase 1 in the Molting and Metamorphosis of the Cigarette Beetle Lasioderma serricorne. International Journal of Molecular Sciences. 2020; 21(7):2449. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072449
Chicago/Turabian StyleYang, Wen-Jia, Kang-Kang Xu, Yi Yan, Can Li, and Dao-Chao Jin. 2020. "Role of Chitin Deacetylase 1 in the Molting and Metamorphosis of the Cigarette Beetle Lasioderma serricorne" International Journal of Molecular Sciences 21, no. 7: 2449. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072449