Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus
Abstract
:1. Introduction
2. Results
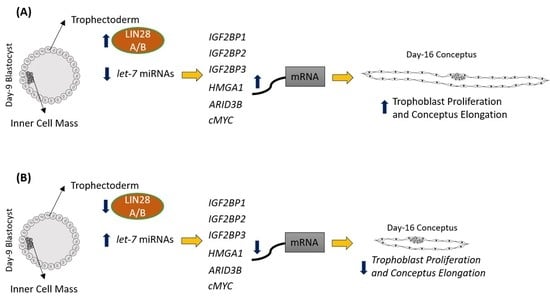
2.1. LIN28 Knockdown in Trophectoderm Resulted in Reduced Proliferation of Trophoblast Cells and Lower Expression of Proliferation-Associated Genes
2.2. Ovine Trophoblast Cells Generated from Day 16 Trophectoderm Had a Significant Reduction in LIN28
2.3. Overexpression of LIN28 in iOTR Cells Resulted in Increased Expression of Proliferation-Associated Genes
2.4. Overexpression of LIN28 Led to Significant Increase in Trophoblast Cell Proliferation
3. Discussion
4. Materials and Methods
4.1. Lentivirus Vector Construction for shRNA Expression
4.2. Lentivirus Vector Construction for Overexpression of LIN28A and LIN28B
4.3. Lentiviral Vector for Immortalizing Passaged Ovine Trophoblast Cells
4.4. Production of Lentiviral Particles
4.5. Blastocyst Collection and Transfer
4.6. Tissue Collection
4.7. Cell Lines
4.8. Overexpression of LIN28A and LIN28B
4.9. RNA Extraction and Real-Time PCR
4.10. Protein Extraction and Western Blot
4.11. Cell Proliferation Assay
4.12. Matrigel Invasion Assay
4.13. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OTR | non-immortalized ovine trophoblast cells |
| iOTR | immortalized ovine trophoblast cells |
| TE | trophectoderm |
| SC | scramble control shRNA |
| AKD | knockdown of LIN28A |
| BKD | knockdown of LIN28B |
| EVC | iOTR cells infected with empty lentiviral expression vectors |
| AKI | iOTR cells overexpressing LIN28A |
| BKI | iOTR cells overexpressing LIN28B |
References
- Knöfler, M.; Haider, S.; Saleh, L.; Pollheimer, J.; Gamage, T.K.J.B.; James, J. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell. Mol. Life Sci. 2019, 76, 3479–3496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aplin, J.D. The cell biological basis of human implantation. Baillieres Best Pract. Res. Clin. Obstet. Gynaecol. 2000, 14, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Red-Horse, K.; Zhou, Y.; Genbacev, O.; Prakobphol, A.; Foulk, R.; McMaster, M.; Fisher, S.J. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Investig. 2004, 114, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Brosens, I.; Pijnenborg, R.; Vercruysse, L.; Romero, R. The "Great Obstetrical Syndromes" are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 2011, 204, 193–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jauniaux, E.; Poston, L.; Burton, G.J. Placental-related diseases of pregnancy: involvement of oxidative stress and implications in human evolution. Hum. Reprod. Update 2006, 12, 747–755. [Google Scholar] [CrossRef] [Green Version]
- Spencer, T.E.; Johnson, G.A.; Bazer, F.W.; Burghardt, R.C. Implantation mechanisms: insights from the sheep. Reprod. Camb. Engl. 2004, 128, 657–668. [Google Scholar] [CrossRef]
- Rowson, L.E.; Moor, R.M. Development of the sheep conceptus during the first fourteen days. J. Anat. 1966, 100, 777–785. [Google Scholar]
- Wintenberger-Torrés, S.; Fléchon, J.E. Ultrastructural evolution of the trophoblast cells of the pre-implantation sheep blastocyst from day 8 to day 18. J. Anat. 1974, 118, 143–153. [Google Scholar]
- Wang, J.; Guillomot, M.; Hue, I. Cellular organization of the trophoblastic epithelium in elongating conceptuses of ruminants. C. R. Biol. 2009, 332, 986–997. [Google Scholar] [CrossRef]
- Spencer, T.E.; Hansen, T.R. Implantation and Establishment of Pregnancy in Ruminants. Adv. Anat. Embryol. Cell Biol. 2015, 216, 105–135. [Google Scholar]
- Shorten, P.R.; Ledgard, A.M.; Donnison, M.; Pfeffer, P.L.; McDonald, R.M.; Berg, D.K. A mathematical model of the interaction between bovine blastocyst developmental stage and progesterone-stimulated uterine factors on differential embryonic development observed on Day 15 of gestation. J. Dairy Sci. 2018, 101, 736–751. [Google Scholar] [CrossRef] [PubMed]
- Farin, C.E.; Imakawa, K.; Hansen, T.R.; McDonnell, J.J.; Murphy, C.N.; Farin, P.W.; Roberts, R.M. Expression of trophoblastic interferon genes in sheep and cattle. Biol. Reprod. 1990, 43, 210–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thatcher, W.W.; Guzeloglu, A.; Mattos, R.; Binelli, M.; Hansen, T.R.; Pru, J.K. Uterine-conceptus interactions and reproductive failure in cattle. Theriogenology 2001, 56, 1435–1450. [Google Scholar] [CrossRef]
- Roberts, R.M.; Chen, Y.; Ezashi, T.; Walker, A.M. Interferons and the maternal-conceptus dialog in mammals. Semin. Cell Dev. Biol. 2008, 19, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Aires, M.B.; Degaki, K.Y.; Dantzer, V.; Yamada, A.T. Bovine placentome development during early pregnancy. Microscope 2014, 1, 390–396. [Google Scholar]
- Lopez-Tello, J.; Arias-Alvarez, M.; Gonzalez-Bulnes, A.; Sferuzzi-Perri, A.N. Models of Intrauterine growth restriction and fetal programming in rabbits. Mol. Reprod. Dev. 2019, 86, 1781–1809. [Google Scholar] [CrossRef] [Green Version]
- Devor, E.J.; Reyes, H.D.; Santillan, D.A.; Santillan, M.K.; Onukwugha, C.; Goodheart, M.J.; Leslie, K.K. Placenta-Specific Protein 1: A Potential Key to Many Oncofetal-Placental OB/GYN Research Questions. Obstet. Gynecol. Int. 2014, 2014, 678984. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.; Anthony, R.V.; Bouma, G.J.; Winger, Q.A. LIN28-let-7 axis regulates genes in immortalized human trophoblast cells by targeting the ARID3B-complex. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 12348–12363. [Google Scholar] [CrossRef] [Green Version]
- West, R.C.; McWhorter, E.S.; Ali, A.; Goetzman, L.N.; Russ, J.E.; Anthony, R.V.; Bouma, G.J.; Winger, Q.A. HMGA2 is regulated by LIN28 and BRCA1 in human placental cells. Biol. Reprod. 2019, 100, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Seabrook, J.L.; Cantlon, J.D.; Cooney, A.J.; McWhorter, E.E.; Fromme, B.A.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Role of LIN28A in Mouse and Human Trophoblast Cell Differentiation. Biol. Reprod. 2013, 89, 95. [Google Scholar] [CrossRef]
- Moss, E.G.; Tang, L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003, 258, 432–442. [Google Scholar] [CrossRef]
- Chan, H.W.; Lappas, M.; Yee, S.W.Y.; Vaswani, K.; Mitchell, M.D.; Rice, G.E. The expression of the let-7 miRNAs and Lin28 signalling pathway in human term gestational tissues. Placenta 2013, 34, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Neumeister, V.; Ma, W.; Xu, J.; Lu, L.; Bordeaux, J.; Maihle, N.J.; Rimm, D.L.; Huang, Y. Lin28 regulates HER2 and promotes malignancy through multiple mechanisms. Cell Cycle 2012, 11, 2486–2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Ma, J.; Xu, J.; Qiao, C.; Branscum, A.; Cardenas, A.; Baron, A.T.; Schwartz, P.; Maihle, N.J.; Huang, Y. Lin28 regulates BMP4 and functions with Oct4 to affect ovarian tumor microenvironment. Cell Cycle 2013, 12, 88–97. [Google Scholar] [CrossRef]
- Hagan, J.P.; Piskounova, E.; Gregory, R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009, 16, 1021–1025. [Google Scholar] [CrossRef] [Green Version]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V.N. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.-K.; Ha, M.; Yoon, M.-J.; Cho, J.; Yeom, K.-H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [Green Version]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; LaPierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R.I. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef] [Green Version]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008, 10, 987–993. [Google Scholar] [CrossRef]
- Newman, M.A.; Thomson, J.M.; Hammond, S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA N. Y. 2008, 14, 1539–1549. [Google Scholar] [CrossRef] [Green Version]
- Ibarra, I.; Erlich, Y.; Muthuswamy, S.K.; Sachidanandam, R.; Hannon, G.J. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007, 21, 3238–3243. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A MicroRNA Regulator with a Macro Role. Cell 2010, 140, 445–449. [Google Scholar] [CrossRef] [Green Version]
- Liao, T.-T.; Hsu, W.-H.; Ho, C.-H.; Hwang, W.-L.; Lan, H.-Y.; Lo, T.; Chang, C.-C.; Tai, S.-K.; Yang, M.-H. let-7 Modulates Chromatin Configuration and Target Gene Repression through Regulation of the ARID3B Complex. Cell Rep. 2016, 14, 520–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.M.; Qian, Z.R.; Wang, E.L.; Sultana, R.; Kudo, E.; Nakasono, M.; Hayashi, T.; Kakiuchi, S.; Sano, T. Frequent overexpression of HMGA1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let-7 downregulation. Br. J. Cancer 2009, 100, 501–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Cai, H.; Liang, Y.; Chen, L.; Wang, X.; Si, R.; Qu, K.; Jiang, Z.; Ma, B.; Miao, C.; et al. Inhibition of c-Myc by let-7b mimic reverses mutidrug resistance in gastric cancer cells. Oncol. Rep. 2015, 33, 1723–1730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchikura, Y.; Matsubara, K.; Matsubara, Y.; Mori, M. P34. Role of high-mobility group A1 protein in trophoblast invasion. Pregnancy Hypertens. Int. J. Womens Cardiovasc. Health 2015, 5, 243. [Google Scholar] [CrossRef]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA Let-7a Down-regulates MYC and Reverts MYC-Induced Growth in Burkitt Lymphoma Cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef] [Green Version]
- Boyerinas, B.; Park, S.-M.; Shomron, N.; Hedegaard, M.M.; Vinther, J.; Andersen, J.S.; Feig, C.; Xu, J.; Burge, C.B.; Peter, M.E. Identification of Let-7–Regulated Oncofetal Genes. Cancer Res. 2008, 68, 2587–2591. [Google Scholar] [CrossRef] [Green Version]
- Bell, J.L.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 2013, 70, 2657–2675. [Google Scholar] [CrossRef] [Green Version]
- JnBaptiste, C.K.; Gurtan, A.M.; Thai, K.K.; Lu, V.; Bhutkar, A.; Su, M.-J.; Rotem, A.; Jacks, T.; Sharp, P.A. Dicer loss and recovery induce an oncogenic switch driven by transcriptional activation of the oncofetal Imp1–3 family. Genes Dev. 2017, 31, 674–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degrauwe, N.; Suvà, M.-L.; Janiszewska, M.; Riggi, N.; Stamenkovic, I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016, 30, 2459–2474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Lu, D.; Liu, L.; Cai, J.; Zhou, Y.; Yang, Y.; Zhang, Y.; Zhang, J. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) promotes lung tumorigenesis via attenuating p53 stability. Oncotarget 2017, 8, 93672–93687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Francia, G.; Zhang, J.-Y. p62/IMP2 stimulates cell migration and reduces cell adhesion in breast cancer. Oncotarget 2015, 6, 32656–32668. [Google Scholar] [CrossRef] [Green Version]
- Mahapatra, L.; Andruska, N.; Mao, C.; Le, J.; Shapiro, D.J. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl. Oncol. 2017, 10, 818–827. [Google Scholar] [CrossRef]
- Zhou, Y.; Meng, X.; Chen, S.; Li, W.; Li, D.; Singer, R.; Gu, W. IMP1 regulates UCA1-mediated cell invasion through facilitating UCA1 decay and decreasing the sponge effect of UCA1 for miR-122-5p. Breast Cancer Res. BCR 2018, 20, 32. [Google Scholar] [CrossRef]
- Schmiedel, D.; Tai, J.; Yamin, R.; Berhani, O.; Bauman, Y.; Mandelboim, O. The RNA binding protein IMP3 facilitates tumor immune escape by downregulating the stress-induced ligands ULPB2 and MICB. eLife 2016, 5, e13426. [Google Scholar] [CrossRef]
- Lederer, M.; Bley, N.; Schleifer, C.; Hüttelmaier, S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin. Cancer Biol. 2014, 29, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Ma, H.; Qi, G.; Chen, F.; Chu, J. Insulin-like growth factor II mRNA-binding protein 3 promotes cell proliferation, migration and invasion in human glioblastoma. Onco. Targets Ther. 2019, 12, 3661–3670. [Google Scholar] [CrossRef] [Green Version]
- Wan, B.-S.; Cheng, M.; Zhang, L. Insulin-like growth factor 2 mRNA-binding protein 1 promotes cell proliferation via activation of AKT and is directly targeted by microRNA-494 in pancreatic cancer. World J. Gastroenterol. 2019, 25, 6063–6076. [Google Scholar] [CrossRef]
- Cao, J.; Mu, Q.; Huang, H. The Roles of Insulin-Like Growth Factor 2 mRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Available online: https://www.hindawi.com/journals/sci/2018/4217259/ (accessed on 18 November 2019).
- Brooks, K.; Burns, G.W.; Moraes, J.G.N.; Spencer, T.E. Analysis of the Uterine Epithelial and Conceptus Transcriptome and Luminal Fluid Proteome During the Peri-Implantation Period of Pregnancy in Sheep. Biol. Reprod. 2016, 95, 88. [Google Scholar] [CrossRef] [PubMed]
- Uchikura, Y.; Matsubara, K.; Muto, Y.; Matsubara, Y.; Fujioka, T.; Matsumoto, T.; Sugiyama, T. Extranuclear Translocation of High-Mobility Group A1 Reduces the Invasion of Extravillous Trophoblasts Involved in the Pathogenesis of Preeclampsia: New Aspect of High-Mobility Group A1. Reprod. Sci. Thousand Oaks Calif. 2017, 24, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Luo, Y.; Tudela, C.; Alexander, J.M.; Mendelson, C.R. The c-Myc-regulated microRNA-17~92 (miR-17~92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol. Cell. Biol. 2013, 33, 1782–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobbs, A.; Gellerman, K.; Hallas, W.M.; Joseph, S.; Yang, C.; Kurkewich, J.; Cowden Dahl, K.D. ARID3B Directly Regulates Ovarian Cancer Promoting Genes. PlLoS ONE 2015, 10, e0131961. [Google Scholar] [CrossRef]
- Roy, L.; Samyesudhas, S.J.; Carrasco, M.; Li, J.; Joseph, S.; Dahl, R.; Cowden Dahl, K.D. ARID3B increases ovarian tumor burden and is associated with a cancer stem cell gene signature. Oncotarget 2014, 5, 8355–8366. [Google Scholar] [CrossRef] [Green Version]
- Ratliff, M.L.; Mishra, M.; Frank, M.B.; Guthridge, J.M.; Webb, C.F. The Transcription Factor ARID3a Is Important for In Vitro Differentiation of Human Hematopoietic Progenitors. J. Immunol. Baltim. Md 1950 2016, 196, 614–623. [Google Scholar] [CrossRef] [Green Version]
- Habir, K.; Aeinehband, S.; Wermeling, F.; Malin, S. A Role for the Transcription Factor Arid3a in Mouse B2 Lymphocyte Expansion and Peritoneal B1a Generation. Front. Immunol. 2017, 8, 1387. [Google Scholar] [CrossRef] [Green Version]
- Rhee, C.; Edwards, M.; Dang, C.; Harris, J.; Brown, M.; Kim, J.; Tucker, H.O. ARID3A is required for mammalian placenta development. Dev. Biol. 2017, 422, 83–91. [Google Scholar] [CrossRef]
- Lala, N.; Girish, G.V.; Cloutier-Bosworth, A.; Lala, P.K. Mechanisms in decorin regulation of vascular endothelial growth factor-induced human trophoblast migration and acquisition of endothelial phenotype. Biol. Reprod. 2012, 87, 59. [Google Scholar] [CrossRef]
- Baker, C.M.; Goetzmann, L.N.; Cantlon, J.D.; Jeckel, K.M.; Winger, Q.A.; Anthony, R.V. Development of ovine chorionic somatomammotropin hormone-deficient pregnancies. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 310, R837–R846. [Google Scholar] [CrossRef] [Green Version]
- Jeckel, K.J.; Boyarko, A.C.; Bouma, G.J.; Winger, Q.A.; Anthony, R.V. Chorionic Somatomammotropin Impacts Early Fetal Growth and Placental Gene Expression. J. Endocrinol. 2018, 237, 301–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purcell, S.H.; Cantlon, J.D.; Wright, C.D.; Henkes, L.E.; Seidel, G.E.; Anthony, R.V. The Involvement of Proline-Rich 15 in Early Conceptus Development in Sheep. Biol. Reprod. 2009, 81, 1112–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsialikas, J.; Romer-Seibert, J. LIN28: roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.-L.; Chen, P.-S.; Johansson, G.; Kuo, M.-L. Function and regulation of let-7 family microRNAs. MicroRNA Shariqah United Arab Emir. 2012, 1, 34–39. [Google Scholar] [CrossRef]
- Straszewski-Chavez, S.L.; Abrahams, V.M.; Alvero, A.B.; Aldo, P.B.; Ma, Y.; Guller, S.; Romero, R.; Mor, G. The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta 2009, 30, 939–948. [Google Scholar] [CrossRef] [Green Version]
- Hayer, A.; Shao, L.; Chung, M.; Joubert, L.-M.; Yang, H.W.; Tsai, F.-C.; Bisaria, A.; Betzig, E.; Meyer, T. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol. 2016, 18, 1311–1323. [Google Scholar] [CrossRef]












© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, A.; Stenglein, M.D.; Spencer, T.E.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus. Int. J. Mol. Sci. 2020, 21, 2549. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072549
Ali A, Stenglein MD, Spencer TE, Bouma GJ, Anthony RV, Winger QA. Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus. International Journal of Molecular Sciences. 2020; 21(7):2549. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072549
Chicago/Turabian StyleAli, Asghar, Mark D. Stenglein, Thomas E. Spencer, Gerrit J. Bouma, Russell V. Anthony, and Quinton A. Winger. 2020. "Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus" International Journal of Molecular Sciences 21, no. 7: 2549. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072549