Differential ESR1 Promoter Methylation in the Peripheral Blood—Findings from the Women 40+ Healthy Aging Study
Abstract
:1. Introduction
2. Results
2.1. Description of Demographic and Biological Measures
2.2. ESR1 Promoter Methylation and Menopausal Status
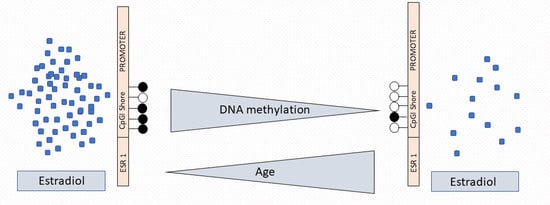
2.3. ESR1 Promoter Methylation, Estradiol Levels, and Age
2.4. CpGI Shore Methylation and Estradiol Levels Among Menopausal Groups
3. Discussion
Strengths and Limitations
4. Materials and Methods
4.1. Subjects
4.2. Biological Sampling
4.3. Methylation Analysis
4.4. Estradiol Measurement
4.5. Procedure and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| E2 | Estradiol |
| ERα | Estrogen receptor α |
| ERβ | Estrogen receptor β |
| GPER | G-protein coupled estrogen receptor |
| ESR1 | ERα gene |
| CpG | Cytosine-guanine dinucleotide |
| CpGI | CpG island |
| STRAW | Stages of Reproductive Aging Workshop |
| DBS | Dried blood spot |
References
- World Health Organization. Life Expectancy. Available online: https://www.who.int/gho/mortality_burden_disease/life_tables/situation_trends_text/en/ (accessed on 1 April 2020).
- Brown, G.C. Living too long. EMBO Rep. 2015, 16, 137–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostan, R.; Monti, D.; Gueresi, P.; Bussolotto, M.; Franceschi, C.; Baggio, G. Gender, aging and longevity in humans: An update of an intriguing/neglected scenario paving the way to a gender-specific medicine. Clin. Sci. 2016, 130, 1711–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Women, Ageing and Health: A Framework for Action: Focus on Gender; World Health Organization: Geneva, Switzerland, 2007. [Google Scholar]
- Horstman, A.M.; Dillon, E.L.; Urban, R.J.; Sheffield-Moore, M. The Role of Androgens and Estrogens on Healthy Aging and Longevity. J. Gerontol. Ser. A 2012, 67, 1140–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiacco, S.; Walther, A.; Ehlert, U. Steroid secretion in healthy aging. Psychoneuroendocrinology 2018. [Google Scholar] [CrossRef]
- Vrtačnik, P.; Ostanek, B.; Mencej-Bedrač, S.; Marc, J. The many faces of estrogen signaling. Biochem. Med. 2014, 24, 329–342. [Google Scholar] [CrossRef] [Green Version]
- Simpson, E.R. Sources of estrogen and their importance. J. Steroid Biochem. Mol. Biol. 2003, 86, 225–230. [Google Scholar] [CrossRef]
- Santoro, N. The menopausal transition. Am. J. Med. 2005, 118, 8–13. [Google Scholar] [CrossRef]
- Gholizadeh, S.; Sadatmahalleh, S.J.; Ziaei, S. The association between estradiol levels and cognitive function in postmenopausal women. Int. J. Reprod. Biomed. 2018, 16, 455. [Google Scholar] [CrossRef]
- Lizcano, F.; Guzmán, G. Estrogen Deficiency and the Origin of Obesity during Menopause. Biomed Res. Int. 2014, 2014, 757461. [Google Scholar] [CrossRef]
- Karvonen-Gutierrez, C.A.; Park, S.K.; Kim, C. Diabetes and Menopause. Curr. Diabetes Rep. 2016, 16, 20. [Google Scholar] [CrossRef]
- Luine, V.N. Estradiol and cognitive function: Past, present and future. Horm. Behav. 2014, 66, 602–618. [Google Scholar] [CrossRef] [Green Version]
- Riecher-Rössler, A.; Kulkarni, J. Estrogens and Gonadal Function in Schizophrenia and Related Psychoses. In Biological Basis of Sex Differences in Psychopharmacology; Neill, J.C., Kulkarni, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 8, pp. 155–171. ISBN 978-3-642-20005-2. [Google Scholar]
- Llaneza, P.; García-Portilla, M.P.; Llaneza-Suárez, D.; Armott, B.; Pérez-López, F.R. Depressive disorders and the menopause transition. Maturitas 2012, 71, 120–130. [Google Scholar] [CrossRef]
- Eastell, R.; O’Neill, T.W.; Hofbauer, L.C.; Langdahl, B.; Reid, I.R.; Gold, D.T.; Cummings, S.R. Postmenopausal osteoporosis. Nat. Rev. Dis. Primers 2016, 2, 16069. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, J.C. A woman’s journey through the reproductive, transitional and postmenopausal periods of life: Impact on cardiovascular and musculo-skeletal risk and the role of estrogen replacement. Maturitas 2011, 70, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Deroo, B.J.; Korach, K.S. Estrogen receptors and human disease. J. Clin. Invest. 2006, 116, 561–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mott, N.N.; Pak, T.R. Estrogen signaling and the aging brain: Context-dependent considerations for postmenopausal hormone therapy. ISRN Endocrinol. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- Wynne, F.L.; Payne, J.A.; Cain, A.E.; Reckelhoff, J.F.; Khalil, R.A. Age-related reduction in estrogen receptor–mediated mechanisms of vascular relaxation in female spontaneously hypertensive rats. Hypertension 2004, 43, 405–412. [Google Scholar] [CrossRef] [Green Version]
- Iorga, A.; Cunningham, C.M.; Moazeni, S.; Ruffenach, G.; Umar, S.; Eghbali, M. The protective role of estrogen and estrogen receptors in cardiovascular disease and the controversial use of estrogen therapy. Biol. Sex Differ. 2017, 8, 33. [Google Scholar] [CrossRef]
- Liu, X.; Shi, H. Regulation of Estrogen Receptor α Expression in the Hypothalamus by Sex Steroids: Implication in the Regulation of Energy Homeostasis. Int. J. Endocrinol. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Paterni, I.; Granchi, C.; Katzenellenbogen, J.A.; Minutolo, F. Estrogen receptors alpha (ERα) and beta (ERβ): Subtype-selective ligands and clinical potential. Steroids 2014, 90, 13–29. [Google Scholar] [CrossRef] [Green Version]
- Heldring, N.; Pike, A.; Andersson, S.; Matthews, J.; Cheng, G.; Hartman, J.; Tujague, M.; Ström, A.; Treuter, E.; Warner, M.; et al. Estrogen Receptors: How Do They Signal and What Are Their Targets. Physiol. Rev. 2007, 87, 905–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piva, R.; Gambari, R.; Zorzato, F.; Kumar, L.; del Senno, L. Analysis of upstream sequences of the human estrogen receptor gene. Biochem. Biophys. Res. Commun. 1992, 183, 996–1002. [Google Scholar] [CrossRef]
- Girgert, R.; Emons, G.; Gründker, C. Estrogen signaling in ERα-negative breast cancer: ERβ and GPER. Front. Endocrinol. 2019, 9, 781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bean, L.A.; Ianov, L.; Foster, T.C. Estrogen receptors, the hippocampus, and memory. Neuroscientist 2014, 20, 534–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouw, A.M.; Efe, G.; Barakat, R.; Preecha, A.; Mehdizadeh, M.; Garan, S.A.; Brooks, G.A. Roles of estrogen receptor-alpha in mediating life span: The hypothalamic deregulation hypothesis. Physiol. Genom. 2017, 49, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.C. Role of estrogen receptor alpha and beta expression and signaling on cognitive function during aging. Hippocampus 2012, 22, 656–669. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Liu, D. Does GPER really function as a G protein-coupled estrogen receptor in vivo? Front. Endocrinol. 2020, 11, 148. [Google Scholar] [CrossRef]
- Hertrampf, T.; Schleipen, B.; Velders, M.; Laudenbach, U.; Fritzemeier, K.H.; Diel, P. Estrogen receptor subtype-specific effects on markers of bone homeostasis. Mol. Cell. Endocrinol. 2008, 291, 104–108. [Google Scholar] [CrossRef]
- Ianov, L.; Kumar, A.; Foster, T.C. Epigenetic regulation of estrogen receptor α contributes to age-related differences in transcription across the hippocampal regions CA1 and CA3. Neurobiol. Aging 2017, 49, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond. Front. Genet. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Mohtat, D.; Susztak, K. Fine tuning gene expression: The epigenome. In Seminars in Nephrology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 30, pp. 468–476. [Google Scholar]
- Baribault, C.; Ehrlich, K.C.; Ponnaluri, V.K.C.; Pradhan, S.; Lacey, M.; Ehrlich, M. Developmentally linked human DNA hypermethylation is associated with down-modulation, repression, and upregulation of transcription. Epigenetics 2018, 13, 275–289. [Google Scholar] [CrossRef]
- Pinzone, J.J.; Stevenson, H.; Strobl, J.S.; Berg, P.E. Molecular and Cellular Determinants of Estrogen Receptor α Expression. Mol. Cell. Biol. 2004, 24, 4605–4612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Ashapkin, V.V.; Kutueva, L.I.; Vanyushin, B.F. Aging as an epigenetic phenomenon. Curr. Genom. 2017, 18, 385–407. [Google Scholar] [CrossRef] [Green Version]
- Avrahami, D.; Li, C.; Zhang, J.; Schug, J.; Avrahami, R.; Rao, S.; Stadler, M.B.; Burger, L.; Schübeler, D.; Glaser, B.; et al. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved β Cell Function. Cell Metab. 2015, 22, 619–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciccarone, F.; Tagliatesta, S.; Caiafa, P.; Zampieri, M. DNA methylation dynamics in aging: How far are we from understanding the mechanisms? Mech. Ageing Dev. 2018, 174, 3–17. [Google Scholar] [CrossRef]
- Flores, K.B.; Wolschin, F.; Amdam, G.V. The role of methylation of DNA in environmental adaptation. Integr. Comp. Biol. 2013, 53, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Tikhodeyev, O.N. Heredity determined by the environment: Lamarckian ideas in modern molecular biology. Sci. Total Environ. 2020, 710, 135521. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Nugent, B.M.; McCarthy, M.M. Developmental and Hormone-Induced Epigenetic Changes to Estrogen and Progesterone Receptor Genes in Brain Are Dynamic across the Life Span. Endocrinology 2010, 151, 4871–4881. [Google Scholar] [CrossRef] [Green Version]
- Tsuboi, K.; Nagatomo, T.; Gohno, T.; Higuchi, T.; Sasaki, S.; Fujiki, N.; Kurosumi, M.; Takei, H.; Yamaguchi, Y.; Niwa, T.; et al. Single CpG site methylation controls estrogen receptor gene transcription and correlates with hormone therapy resistance. J. Steroid Biochem. Mol. Biol. 2017, 171, 209–217. [Google Scholar] [CrossRef]
- Issa, J.-P.J.; Ottaviano, Y.L.; Celano, P.; Hamilton, S.R.; Davidson, N.E.; Baylin, S.B. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat. Genet. 1994, 7, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Post, W.S.; Goldschmidt-Clermont, P.J.; Wilhide, C.C.; Heldman, A.W.; Sussman, M.S.; Ouyang, P.; Milliken, E.E.; Issa, J.-P.J. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc. Res. 1999, 43, 985–991. [Google Scholar] [CrossRef]
- Gaudet, M.M.; Campan, M.; Figueroa, J.D.; Yang, X.R.; Lissowska, J.; Peplonska, B.; Brinton, L.A.; Rimm, D.L.; Laird, P.W.; Garcia-Closas, M.; et al. DNA Hypermethylation of ESR1 and PGR in Breast Cancer: Pathologic and Epidemiologic Associations. Cancer Epidemiol. Biomark. Prev. 2009, 18, 3036–3043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirn, V.; Strake, L.; Thangarajah, F.; Richters, L.; Eischeid, H.; Koitzsch, U.; Odenthal, M.; Fries, J. ESR1-promoter-methylation status in primary breast cancer and its corresponding metastases. Clin. Exp. Metastasis 2018, 35, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Guo, Y.; Lu, Y. Prognostic role of methylated GSTP1, p16, ESR1 and PITX2 in patients with breast cancer: A systematic meta-analysis under the guideline of PRISMA. Medicine 2017, 96, e7476. [Google Scholar] [CrossRef]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database J. Biol. Databases Curation 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Pennacchio, L.A.; Bickmore, W.; Dean, A.; Nobrega, M.A.; Bejerano, G. Enhancers: Five essential questions. Nat. Rev. Genet. 2013, 14, 288–295. [Google Scholar] [CrossRef]
- Bell, E.; Curry, E.W.; Megchelenbrink, W.; Jouneau, L.; Brochard, V.; Tomaz, R.A.; Mau, K.H.T.; Atlasi, Y.; de Souza, R.A.; Marks, H. Dynamic CpG methylation delineates subregions within super-enhancers selectively decommissioned at the exit from naive pluripotency. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Lin, X.; Li, L.; Liu, X.; Tian, J.; Zheng, W.; Li, J.; Wang, L. Genome-wide analysis of aberrant methylation of enhancer DNA in human osteoarthritis. BMC Med. Genom. 2020, 13. [Google Scholar] [CrossRef]
- Stadler, M.B.; Murr, R.; Burger, L.; Ivanek, R.; Lienert, F.; Schöler, A.; van Nimwegen, E.; Wirbelauer, C.; Oakeley, E.J.; Gaidatzis, D.; et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011, 480, 490–495. [Google Scholar] [CrossRef]
- Magnusson, M.; Lu, E.X.; Larsson, P.; Ulfhammer, E.; Bergh, N.; Carén, H.; Jern, S. Dynamic Enhancer Methylation - A Previously Unrecognized Switch for Tissue-Type Plasminogen Activator Expression. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Field, A.E.; Robertson, N.A.; Wang, T.; Havas, A.; Ideker, T.; Adams, P.D. DNA Methylation Clocks in Aging: Categories, Causes, and Consequences. Mol. Cell 2018, 71, 882–895. [Google Scholar] [CrossRef] [Green Version]
- Jylhävä, J.; Pedersen, N.L.; Hägg, S. Biological age predictors. EBioMedicine 2017, 21, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabian, D.; Flatt, T. The evolution of aging. Nat. Educ. Knowl. 2011, 3, 1–10. [Google Scholar]
- Brinton, R.D.; Yao, J.; Yin, F.; Mack, W.J.; Cadenas, E. Perimenopause as a neurological transition state. Nat. Rev. Endocrinol. 2015, 11, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Kreft, Ł.; Soete, A.; Hulpiau, P.; Botzki, A.; Saeys, Y.; De Bleser, P. ConTra v3: A tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res. 2017, 45, W490–W494. [Google Scholar] [CrossRef] [PubMed]
- Champagne, F.A.; Weaver, I.C.G.; Diorio, J.; Dymov, S.; Szyf, M.; Meaney, M.J. Maternal Care Associated with Methylation of the Estrogen Receptor-α1b Promoter and Estrogen Receptor-α Expression in the Medial Preoptic Area of Female Offspring. Endocrinology 2006, 147, 2909–2915. [Google Scholar] [CrossRef]
- Héberlé, É.; Bardet, A.F. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019, 63, 727–741. [Google Scholar] [CrossRef] [Green Version]
- Campesi, I.; Sanna, M.; Zinellu, A.; Carru, C.; Rubattu, L.; Bulzomi, P.; Seghieri, G.; Tonolo, G.; Palermo, M.; Rosano, G.; et al. Oral contraceptives modify DNA methylation and monocyte-derived macrophage function. Biol. Sex Differ. 2012, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Ho, S.-M. Epigenetics meets endocrinology. J. Mol. Endocrinol. 2011, 46, R11–R32. [Google Scholar] [CrossRef] [Green Version]
- Hannon, E.; Lunnon, K.; Schalkwyk, L.; Mill, J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 2015, 10, 1024–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.K.; Kilaru, V.; Kocak, M.; Almli, L.M.; Mercer, K.B.; Ressler, K.J.; Tylavsky, F.A.; Conneely, K.N. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genom. 2014, 15, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wilson, R.; Heiss, J.; Breitling, L.P.; Saum, K.-U.; Schöttker, B.; Holleczek, B.; Waldenberger, M.; Peters, A.; Brenner, H. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun. 2017, 8, 14617. [Google Scholar] [CrossRef] [PubMed]
- Khakpour, G.; Pooladi, A.; Izadi, P.; Noruzinia, M.; Bazzaz, J.T. DNA methylation as a promising landscape: A simple blood test for breast cancer prediction. Tumor Biol. 2015, 36, 4905–4912. [Google Scholar] [CrossRef]
- Suga, Y.; Miyajima, K.; Oikawa, T.; Maeda, J.; Usuda, J.; Kajiwara, N.; Ohira, T.; Uchida, O.; Tsuboi, M.; Hirano, T. Quantitative p16 and ESR1 methylation in the peripheral blood of patients with non-small cell lung cancer. Oncol. Rep. 2008, 20, 1137–1142. [Google Scholar]
- Martínez-Galán, J.; Torres-Torres, B.; Núñez, M.I.; López-Peñalver, J.; Del Moral, R.; Ruiz De Almodóvar, J.M.; Menjón, S.; Concha, Á.; Chamorro, C.; Ríos, S.; et al. ESR1 gene promoter region methylation in free circulating DNA and its correlation with estrogen receptor protein expression in tumor tissue in breast cancer patients. BMC Cancer 2014, 14, 59. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, A.; Sapehia, D.; Thakur, S.; Mohanraj, P.S.; Bagga, R.; Kaur, J. Effect of imbalance in folate and vitamin B12 in maternal/parental diet on global methylation and regulatory miRNAs. Sci. Rep. 2019, 9, 17602. [Google Scholar] [CrossRef]
- Mernone, L.; Fiacco, S.; Ehlert, U. Psychobiological Factors of Sexual Functioning in Aging Women – Findings From the Women 40+ Healthy Aging Study. Front. Psychol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Fiacco, S.; Gardini, E.S.; Mernone, L.; Schick, L.; Ehlert, U. Table_1_DNA Methylation in Healthy Older Adults With a History of Childhood Adversity—Findings From the Women 40+ Healthy Aging Study. Front. Psychiatry 2019, 10, 777. [Google Scholar] [CrossRef] [Green Version]
- Fiacco, S.; Mernone, L.; Ehlert, U. Psychobiological indicators of the subjectively experienced health status-findings from the Women 40+ Healthy Aging Study. BMC Womens. Health 2020, 20, 16. [Google Scholar] [CrossRef]
- Harlow, S.D.; Gass, M.; Hall, J.E.; Lobo, R.; Maki, P.; Rebar, R.W.; Sherman, S.; Sluss, P.M.; de Villiers, T.J.; Group, S. + 10 C. Executive summary of the Stages of Reproductive Aging Workshop + 10: Addressing the unfinished agenda of staging reproductive aging. J. Clin. Endocrinol. Metab. 2012, 97, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.G.; Carr, B.R. The Normal Menstrual Cycle and the Control of Ovulation. In Endotext; De Groot, L.J., Chrousos, G., Dungan, K., Feingold, K.R., Grossman, A., Hershman, J.M., Koch, C., Korbonits, M., McLachlan, R., New, M., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Kim, S.O.O.W.; Kim, N.N.; Jeong, S.-J.; Munarriz, R.; Goldstein, I.; Traish, A.M. Modulation of rat vaginal blood flow and estrogen receptor by estradiol. J. Urol. 2004, 172, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Aberg, K.A.; Xie, L.Y.; Nerella, S.; Copeland, W.E.; Costello, E.J.; van den Oord, E.J.C.G. High quality methylome-wide investigations through next-generation sequencing of DNA from a single archived dry blood spot. Epigenetics 2013, 8, 542–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, S.; Obrist, R.; Ehlert, U. How and when to use dried blood spots in psychoneuroendocrinological research. Psychoneuroendocrinology 2019, 108, 190–196. [Google Scholar] [CrossRef]
- Chen, G.G.; Gross, J.A.; Lutz, P.-E.; Vaillancourt, K.; Maussion, G.; Bramoulle, A.; Théroux, J.-F.; Gardini, E.S.; Ehlert, U.; Bourret, G.; et al. Medium throughput bisulfite sequencing for accurate detection of 5-methylcytosine and 5-hydroxymethylcytosine. BMC Genom. 2017, 18. [Google Scholar] [CrossRef] [Green Version]
- Li, L.-C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, J.L.; Kelly, R.P.; Lowell, N.C.; Port, J.A. Indexed PCR primers induce template-specific bias in large-scale DNA sequencing studies. PLoS ONE 2016, 11, e0148698. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y.; et al. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [Green Version]
- Dielen, C.; Fiers, T.; Somers, S.; Deschepper, E.; Gerris, J. Correlation between saliva and serum concentrations of estradiol in women undergoing ovarian hyperstimulation with gonadotropins for IVF/ICSI. Facts, Views Vis. ObGyn 2019, 9, 85–91. [Google Scholar]
- Wu, C.-H.; Motohashi, T.; Abdel-Rahman, H.A.; Flickinger, G.L.; Mikhail, G. Free and Protein-Bound Plasma Estradiol-17β During the Menstrual Cycle. J. Clin. Endocrinol. Metab. 1976, 43, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Package ‘Robustbase’. Available online: http://robustbase.r-forge.r-project.org/ (accessed on 11 January 2019).
- Team, R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- Sullivan, G.M.; Feinn, R. Using effect size—Or why the P value is not enough. J. Grad. Med. Educ. 2012, 4, 279–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| All | PRE | PERI | POST | |
|---|---|---|---|---|
| n | 130 | 51 | 16 | 63 |
| E2 (pmol/L) (mean/SD) | 5.6/4.5 | 8.2/4.7 | 5.7/4.5 | 3.7/3.2 |
| Age (y) (mean/median/range) | 53.2/52.5/40–73 | 45.1/45/40–57 | 51.4/51/47–56 | 60.2/59/50–73 |
| ESR1 CpGI shore methylation (%) (mean/SD) | 76.7/12 | 79.3/8.4 | 78.1/9.2 | 74.4/14.7 |
| ESR1 promoter B methylation (%) (mean/SD) | 3.5/3.14 | 3.2/2 | 3.9/3.3 | 3.7/3.8 |
| ESR1 promoter A methylation (%) (mean/SD) | 3.9/3.3 | 3.7/2.5 | 5/4 | 3.7/3.7 |
| E2 | β | p | Age | β | p | |
|---|---|---|---|---|---|---|
| CpG 1 | 0.64 | 0.097 | −0.25 | 0.599 | ||
| CpG 2 | 0.12 | 0.733 | −0.42 | 0.154 | ||
| CpG 3 | 0.54 | 0.018 | 0.09 | 0.618 | ||
| CpG 4 | −0.01 | 0.978 | −0.00 | 0.989 | ||
| CpG 5 | 0.42 | 0.051 | 0.09 | 0.661 | ||
| CpG 6 | 0.13 | 0.587 | 0.14 | 0.293 | ||
| CpG 7 | 0.28 | 0.100 | 0.04 | 0.831 | ||
| CpG 8 | −0.06 | 0.797 | −0.28 | 0.422 | ||
| CpG 9 | 0.65 | 0.019 | −0.33 | 0.419 |
| Target | Forward Primer | Reverse Primer | GRC h37 (hg19) | T (°C) |
|---|---|---|---|---|
| CpGI shore | ACACTGACGACATGGTTCTACA NNN GTTTTTTGTGAGTAGATAGTAAGTT | TACGGTAGCAGAGACTTGGTCT NNN AAACCTACCCTACTAAATCAAAAAC | chr6: 152126660–152126963 | 58 |
| Promoter B | ACACTGACGACATGGTTCTACA NNN GGGGAATTAAATAGAAAGAGAGATAAATAG | TACGGTAGCAGAGACTTGGTCT NNN CCAAAAAACAACTTCCCTAAACTT | chr6: 152128433–152128671 | 60 |
| Promoter A | ACACTGACGACATGGTTCTACA NNN AGATTAGTATTTAAAGTTGGAGGTT | TACGGTAGCAGAGACTTGGTCT NNN ATATAAAAAATCATAATCATAATCC | chr6: 152128780–152129067 | 54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gardini, E.S.; Chen, G.G.; Fiacco, S.; Mernone, L.; Willi, J.; Turecki, G.; Ehlert, U. Differential ESR1 Promoter Methylation in the Peripheral Blood—Findings from the Women 40+ Healthy Aging Study. Int. J. Mol. Sci. 2020, 21, 3654. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103654
Gardini ES, Chen GG, Fiacco S, Mernone L, Willi J, Turecki G, Ehlert U. Differential ESR1 Promoter Methylation in the Peripheral Blood—Findings from the Women 40+ Healthy Aging Study. International Journal of Molecular Sciences. 2020; 21(10):3654. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103654
Chicago/Turabian StyleGardini, Elena S., Gary G. Chen, Serena Fiacco, Laura Mernone, Jasmine Willi, Gustavo Turecki, and Ulrike Ehlert. 2020. "Differential ESR1 Promoter Methylation in the Peripheral Blood—Findings from the Women 40+ Healthy Aging Study" International Journal of Molecular Sciences 21, no. 10: 3654. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103654