Evidence for Enhanced Exosome Production in Aromatase Inhibitor-Resistant Breast Cancer Cells
Abstract
:1. Introduction
2. Results
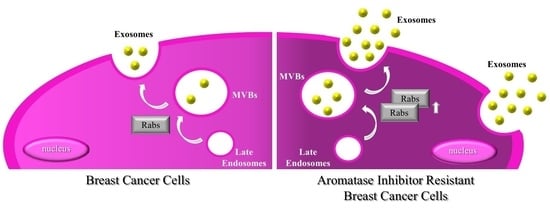
2.1. MCF-7 LTED (Long-Term Estrogen Deprived) Cells Exhibit Increased Exosome Production
2.2. Quantitative Proteomic Analysis Shows Extensive Changes of Protein Expression in MCF-7 LTED Cells Compared with MCF-7 Cells
2.3. MCF-7 LTED Cells Show Enhanced Rab GTPase Protein Expression
3. Discussion
4. Materials and Methods
4.1. Antibodies
4.2. Cell Cultures
4.3. Isolation of Tumor-Derived Exosomes
4.4. Transmission Electron Microscopy (TEM)
4.5. Immunoblot Analysis
4.6. Nanoparticle Tracking Analysis (NTA)
4.7. RNA Extraction and Real-Time RT-PCR Assays
4.8. TUNEL Assay
4.9. Proteomic Analysis
4.9.1. Protein Digestion for MS Analysis
4.9.2. Mass Spectrometry Analysis
4.9.3. MS Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Murphy, C.G.; Dickler, M.N. Endocrine resistance in hormone-responsive breast cancer: Mechanisms and therapeutic strategies. Endocr. Relat. Cancer 2016, 23, R337–R352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, L.-A.; Farmer, I.; Johnston, S.R.D.; Ali, S.; Dowsett, M. Elevated ERK1/ERK2/estrogen receptor cross-talk enhances estrogen-mediated signaling during long-term estrogen deprivation. Endocr. Relat. Cancer 2005, 12, S75–S84. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, G.J.; Jelovac, D.; Long, B.; Brodie, A. The Role of Growth Factor Receptor Pathways in Human Breast Cancer Cells Adapted to Long-term Estrogen Deprivation. Cancer Res. 2005, 65, 3903–3910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santen, R.J.; Song, R.X.; Zhang, Z.; Kumar, R.; Jeng, M.H.; Masamura, A.; Lawrence, J., Jr.; Berstein, L.; Yue, W. Long-term estradiol deprivation in breast cancer cells up-regulates growth factor signaling and enhances estrogen sensitivity. Endocr. Relat. Cancer 2005, 12, S61–S73. [Google Scholar] [CrossRef] [PubMed]
- Barone, I.; Iacopetta, D.; Covington, K.R.; Cui, Y.; Tsimelzon, A.; Beyer, A.; Andò, S.; Fuqua, S.A.W.; Anna, T. Phosphorylation of the mutant K303R estrogen receptor α at serine 305 affects aromatase inhibitor sensitivity. Oncogene 2010, 29, 2404–2414. [Google Scholar] [CrossRef] [Green Version]
- Chen, S. An “Omics” Approach to Determine the Mechanisms of Acquired Aromatase Inhibitor Resistance. Omics A J. Integr. Biol. 2011, 15, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Thangavel, C.; Dean, J.L.; Ertel, A.; Knudsen, K.E.; Aldaz, C.M.; Witkiewicz, A.K.; Clarke, R.; Knudsen, E.S. Therapeutically activating RB: Reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr. Relat. Cancer 2011, 18, 333–345. [Google Scholar] [CrossRef]
- Dittmer, J.; Leyh, B. The impact of tumor stroma on drug response in breast cancer. Semin. Cancer Biol. 2015, 31, 3–15. [Google Scholar] [CrossRef]
- Ma, C.X.; Reinert, T.; Chmielewska, I.; Ellis, M.J. Mechanisms of aromatase inhibitor resistance. Nat. Rev. Cancer 2015, 15, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Bennit, H.F.; Turay, D.; Perez, M.; Mirshahidi, S.; Yuan, Y.; Wall, N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176. [Google Scholar] [CrossRef] [Green Version]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharm. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Dutta, S.; Warshall, C.; Bandyopadhyay, C.; Dutta, D.; Chandran, B. Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS ONE 2014, 9, e97580. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.L.; May, L.; Lhotak, V.; Shahrzad, S.; Shirasawa, S.; Weitz, J.I.; Coomber, B.L.; Mackman, N.; Rak, J.W. Oncogenic events regulate tissue factor expression in colorectal cancer cells: Implications for tumor progression and angiogenesis. Blood 2005, 105, 1734–1741. [Google Scholar] [CrossRef]
- Antonyak, M.A.; Li, B.; Boroughs, L.K.; Johnson, J.L.; Druso, J.E.; Bryant, K.L.; Holowka, D.A.; Cerione, R.A. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4852–4857. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Chow, A.; Zhou, W.; Liu, L.; Fong, M.Y.; Champer, J.; Van Haute, D.; Chin, A.R.; Ren, X.; Gugiu, B.G.; Meng, Z.; et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-κB. Sci. Rep. 2014, 4, 5750. [Google Scholar] [CrossRef] [Green Version]
- Kenific, C.M.; Nogués, L.; Lyden, D. Pre-Metastatic Niche Formation Has Taken Its TOLL. Cancer Cell 2016, 30, 189–191. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.A.; Park, H.; Lim, E.H.; Lee, K.W. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int. J. Oncol. 2011, 40, 130–138. [Google Scholar] [CrossRef] [Green Version]
- Gutkin, A.; Uziel, O.; Beery, E.; Nordenberg, J.; Pinchasi, M.; Goldvaser, H.; Henick, S.; Goldberg, M.; Lahav, M. Tumor cells derived exosomes contain hTERT mRNA and transform nonmalignant fibroblasts into telomerase positive cells. Oncotarget 2016, 7, 59173–59188. [Google Scholar] [CrossRef] [Green Version]
- Lv, M.M.; Zhu, X.Y.; Chen, W.X.; Zhong, S.L.; Hu, Q.; Ma, T.F.; Zhang, J.; Chen, L.; Tang, J.H.; Zhao, J.H. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumor Biol 2014, 35, 10773–10779. [Google Scholar] [CrossRef]
- Martinez, V.G.; O’Neill, S.; Salimu, J.; Breslin, S.; Clayton, A.; Crown, J.; O’Driscoll, L. Resistance to HER2-targeted anti-cancer drugs is associated with immune evasion in cancer cells and their derived extracellular vesicles. Oncoimmunology 2017, 6, e1362530. [Google Scholar] [CrossRef]
- Dong, H.; Wang, W.; Chen, R.; Zhang, Y.; Zou, K.; Ye, M.; He, X.; Zhang, F.; Han, J. Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 2018, 53, 1013–1026. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Hu, J.; Lu, P.; Cao, H.; Yu, C.; Li, X.; Qian, X.; Yang, X.; Yang, Y.; Han, N.; et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 43. [Google Scholar] [CrossRef]
- Del Re, M.; Bertolini, I.; Crucitta, S.; Fontanelli, L.; Rofi, E.; De Angelis, C.; Diodati, L.; Cavallero, D.; Gianfilippo, G.; Salvadori, B.; et al. Overexpression of TK1 and CDK9 in plasma-derived exosomes is associated with clinical resistance to CDK4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Res. Treat. 2019, 178, 57–62. [Google Scholar] [CrossRef]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.G.; Yang, M.F.; Ren, Y.Q.; Wu, C.H.; Wang, L.Q. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharm. Sci. 2016, 20, 4362–4368. [Google Scholar]
- Semina, S.E.; Scherbakov, A.M.; Vnukova, A.A.; Bagrov, D.V.; Evtushenko, E.G.; Safronova, V.M.; Golovina, D.A.; Lyubchenko, L.N.; Gudkova, M.V.; Krasil’nikov, M.A. Exosome-Mediated Transfer of Cancer Cell Resistance to Antiestrogen Drugs. Molecules 2018, 23, 829. [Google Scholar] [CrossRef] [Green Version]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [Green Version]
- Lombardo, Y.; Faronato, M.; Filipovic, A.; Vircillo, V.; Magnani, L.; Coombes, R.C. Nicastrin and Notch4 drive endocrine therapy resistance and epithelial to mesenchymal transition in MCF7 breast cancer cells. Breast Cancer Res. 2014, 16, R62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell. Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Lynch, C.; Panagopoulou, M.; Gregory, C.D. Extracellular Vesicles Arising from Apoptotic Cells in Tumors: Roles in Cancer Pathogenesis and Potential Clinical Applications. Front. Immunol. 2017, 8, 1174. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Leal, J.B.; Seabra, M.C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001, 313, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Blanc, L.; Vidal, M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small Gtpases 2018, 9, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.D.; Zhang, Y.J.; Wei, Y.H.; Cho, J.H.; Morris, L.E.; Wang, H.Y.; Zheng, X.F. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell 2014, 26, 754–769. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.H.; Li, X.X.; Yang, Y.; Zhang, M.Y.; Rao, H.L.; Wang, H.Y.; Zheng, X.F. Aberrant amino acid signaling promotes growth and metastasis of hepatocellular carcinomas through Rab1A-dependent activation of mTORC1 by Rab1A. Oncotarget 2015, 6, 20813–20828. [Google Scholar] [CrossRef] [Green Version]
- Bao, Z.S.; Li, M.Y.; Wang, J.Y.; Zhang, C.B.; Wang, H.J.; Yan, W.; Liu, Y.W.; Zhang, W.; Chen, L.; Jiang, T. Prognostic value of a nine-gene signature in glioma patients based on mRNA expression profiling. CNS Neurosci. Ther. 2014, 20, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Abd Elmageed, Z.Y.; Yang, Y.; Thomas, R.; Ranjan, M.; Mondal, D.; Moroz, K.; Fang, Z.; Rezk, B.M.; Moparty, K.; Sikka, S.C.; et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells 2014, 32, 983–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzeng, H.T.; Wang, Y.C. Rab-mediated vesicle trafficking in cancer. J. Biomed. Sci. 2016, 23, 70. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.L.; Gong, C.; Chen, C.H.; Hu, H.; Huang, P.; Zheng, M.; Yao, Y.; Wei, S.; Wulf, G.; Lieberman, J.; et al. The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via Erk signaling activation. Cell Rep. 2015, 11, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.S.; Yin, P.H.; Tseng, L.M.; Yang, C.H.; Hsu, C.Y.; Lee, M.Y.; Horng, C.F.; Chi, C.W. Rab5A is associated with axillary lymph node metastasis in breast cancer patients. Cancer Sci. 2011, 102, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
- Frittoli, E.; Palamidessi, A.; Marighetti, P.; Confalonieri, S.; Bianchi, F.; Malinverno, C.; Mazzarol, G.; Viale, G.; Martin-Padura, I.; Garre, M.; et al. A RAB5/RAB4 recycling circuitry induces a proteolytic invasive program and promotes tumor dissemination. J. Cell Biol. 2014, 206, 307–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Yan, Y.; Liu, F.; Kang, H.; Xu, F.; Xiao, W.; Wang, H.; Wang, Y. Knockdown of Rab7a suppresses the proliferation, migration, and xenograft tumor growth of breast cancer cells. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.O.; Shin, S.; Mercurio, A.M. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the α6β4 integrin. Cancer Res. 2005, 65, 2761–2769. [Google Scholar] [CrossRef] [Green Version]
- Recchi, C.; Seabra, M.C. Novel functions for Rab GTPases in multiple aspects of tumour progression. Biochem. Soc. Trans. 2012, 40, 1398–1403. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Théry, C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [Green Version]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; Ben David, G.; Shlomi, T.; Gil, Z. Transfer of miRNA in Macrophage-Derived Exosomes Induces Drug Resistance in Pancreatic Adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef] [Green Version]
- Shen, D.W.; Gottesman, M.M. RAB8 enhances TMEM205-mediated cisplatin resistance. Pharm. Res. 2012, 29, 643–650. [Google Scholar] [CrossRef] [Green Version]
- Dorayappan, K.D.P.; Wanner, R.; Wallbillich, J.J.; Saini, U.; Zingarelli, R.; Suarez, A.A.; Cohn, D.E.; Selvendiran, K. Hypoxia-induced exosomes contribute to a more aggressive and chemoresistant ovarian cancer phenotype: A novel mechanism linking STAT3/Rab proteins. Oncogene 2018, 37, 3806–3821. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Qi, R.; Liu, X.; Qian, L.; Wu, Z. Rab18 overexpression promotes proliferation and chemoresistance through regulation of mitochondrial function in human gastric cancer. Onco Targets. Ther. 2018, 11, 7805–7820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giordano, C.; Gelsomino, L.; Barone, I.; Panza, S.; Augimeri, G.; Bonofiglio, D.; Rovito, D.; Naimo, G.D.; Leggio, A.; Catalano, S.; et al. Leptin Modulates Exosome Biogenesis in Breast Cancer Cells: An Additional Mechanism in Cell-to-Cell Communication. J. Clin. Med. 2019, 8, 1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovito, D.; Giordano, C.; Plastina, P.; Barone, I.; De Amicis, F.; Mauro, L.; Rizza, P.; Lanzino, M.; Catalano, S.; Bonofiglio, D.; et al. Omega-3 DHA- and EPA-dopamine conjugates induce PPARγ-dependent breast cancer cell death through autophagy and apoptosis. Biochim. Biophys. Acta BBA Gen. Subj. 2015, 1850, 2185–2195. [Google Scholar] [CrossRef]
- Mauro, L.; Sisci, D.; Bartucci, M.; Salerno, M.; Kim, J.; Tam, T.; Guvakova, M.A.; Ando, S.; Surmacz, E. SHC- α5β1 integrin interactions regulate breast cancer cell adhesion and motility. Exp. Cell Res. 1999, 252, 439–448. [Google Scholar] [CrossRef]
- Catalano, S.; Panza, S.; Malivindi, R.; Giordano, C.; Barone, I.; Bossi, G.; Lanzino, M.; Sirianni, R.; Mauro, L.; Sisci, D.; et al. Inhibition of leydig tumor growth by farnesoid X receptor activation: The in vitro and in vivo basis for a novel therapeutic strategy. Int. J. Cancer 2013, 132, 2237–2247. [Google Scholar] [CrossRef] [PubMed]
- Catalano, S.; Malivindi, R.; Giordano, C.; Gu, G.; Panza, S.; Bonofiglio, D.; Lanzino, M.; Sisci, D.; Panno, M.L.; Andò, S. Farnesoid X receptor, through the binding with steroidogenic factor 1-responsive element, inhibits aromatase expression in tumor Leydig cells. J. Biol. Chem. 2010, 285, 5581–5593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giordano, C.; Rovito, D.; Barone, I.; Mancuso, R.; Bonofiglio, D.; Giordano, F.; Catalano, S.; Gabriele, B.; Andò, S. Benzofuran-2-acetic ester derivatives induce apoptosis in breast cancer cells by upregulating p21Cip/WAF1 gene expression in p53-independent manner. DNA Repair 2017, 51, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Bakunts, A.; Orsi, A.; Vitale, M.; Cattaneo, A.; Lari, F.; Tade, L.; Sitia, R.; Raimondi, A.; Bachi, A.; van Anken, E. Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. Elife 2017, 6. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudas, J.; Skvortsov, S.; Ganswindt, U.; Riechelmann, H.; Skvortsova, I.-I. Therapy resistance mediated by exosomes. Mol. Cancer 2019, 18, 58. [Google Scholar] [CrossRef]




| Term | Count | % | p Value | Fold Enrichment | FDR |
|---|---|---|---|---|---|
| RNA processing | 188 | 7.75 | 1.80 × 10−31 | 2.33 | 3.35 × 10−28 |
| Establishment of protein localization | 235 | 9.69 | 1.47 × 10−30 | 2.07 | 2.73 × 10−27 |
| Protein transport | 233 | 9.60 | 2.45 × 10−30 | 2.07 | 4.56 × 10−27 |
| Translation | 133 | 5.48 | 8.43 × 10−30 | 2.72 | 1.57 × 10−26 |
| Protein localization | 255 | 10.51 | 4.90 × 10−29 | 1.96 | 9.13 × 10−26 |
| RNA splicing | 118 | 4.86 | 5.10 × 10−28 | 2.81 | 9.49 × 10−25 |
| mRNA metabolic processing | 138 | 5.69 | 4.41 × 10−27 | 2.53 | 8.20 × 10−24 |
| Intracellular transport | 203 | 8.38 | 4.87 × 10−27 | 2.09 | 9.07 × 10−24 |
| mRNA processing | 123 | 5.07 | 2.14 × 10−25 | 2.60 | 3.98 × 10−22 |
| Vesicle-mediated transport | 168 | 6.92 | 1.39 × 10−19 | 1.98 | 2.59 × 10−16 |
| Protein ID | Protein Name | Gene Names | Ratio AVG |
|---|---|---|---|
| Q969Q5 | Ras-related protein Rab-24 | RAB24 | 6.41074493 |
| Q6WKZ4 | Rab11 family-interacting protein 1 | RAB11FIP1 | 3.283109519 |
| Q13637 | Ras-related protein Rab-32 | RAB32 | 3.134080381 |
| A0A024R2K1 | Ras-related protein Rab-5A | RAB5A | 2.684524433 |
| Q8WUD1 | Ras-related protein Rab-2B | RAB2B | 2.358702001 |
| A0A024RD41 | Ras-related protein Rab-23 | RAB23 | 1.874367334 |
| Q5HYI8 | Rab-like protein 3 | RABL3 | 1.832400186 |
| Q9UL26 | Ras-related protein Rab-22A | RAB22A | 1.72827705 |
| Q6IQ22 | Ras-related protein Rab-12 | RAB12 | 1.608127936 |
| A0A024RBA9 | Ras-related protein Rab-21 | RAB21 | 1.583370444 |
| A0A024R5J5 | Ras-related protein Rab-6A | RAB6A | 1.470976838 |
| A0A024R7V6 | Ras-related protein Rab-2A | RAB2; RAB2A | 1.455072096 |
| Q9NP72 | Ras-related protein Rab-18 | RAB18 | 1.451684135 |
| A0A024R7G2 | Ras-related protein Rab-3D | RAB3D | 1.42786787 |
| A0A024R7I7 | Ras-related protein Rab-3A | RAB3A | 1.41943675 |
| Q15907 | Ras-related protein Rab-11B; Ras-related protein Rab-11A | RAB11B; RAB11A | 1.403714286 |
| A0A024R845 | Ras-related protein Rab-14 | RAB14 | 1.381289829 |
| A0A024RB09 | Ras-related protein Rab-5B | RAB5B | 1.333981668 |
| A0A158RFU6 | Ras-related protein Rab-7a | RAB7A | 1.332295086 |
| O00194 | Ras-related protein Rab-27B | RAB27B | 1.327797939 |
| A0A024R1U4 | Ras-related protein Rab-5C | RAB5C | 1.307181423 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Augimeri, G.; La Camera, G.; Gelsomino, L.; Giordano, C.; Panza, S.; Sisci, D.; Morelli, C.; Győrffy, B.; Bonofiglio, D.; Andò, S.; et al. Evidence for Enhanced Exosome Production in Aromatase Inhibitor-Resistant Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5841. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165841
Augimeri G, La Camera G, Gelsomino L, Giordano C, Panza S, Sisci D, Morelli C, Győrffy B, Bonofiglio D, Andò S, et al. Evidence for Enhanced Exosome Production in Aromatase Inhibitor-Resistant Breast Cancer Cells. International Journal of Molecular Sciences. 2020; 21(16):5841. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165841
Chicago/Turabian StyleAugimeri, Giuseppina, Giusi La Camera, Luca Gelsomino, Cinzia Giordano, Salvatore Panza, Diego Sisci, Catia Morelli, Balázs Győrffy, Daniela Bonofiglio, Sebastiano Andò, and et al. 2020. "Evidence for Enhanced Exosome Production in Aromatase Inhibitor-Resistant Breast Cancer Cells" International Journal of Molecular Sciences 21, no. 16: 5841. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165841