IKKγ/NEMO Is Required to Confer Antimicrobial Innate Immune Responses in the Yellow Mealworm, Tenebrio Molitor
Abstract
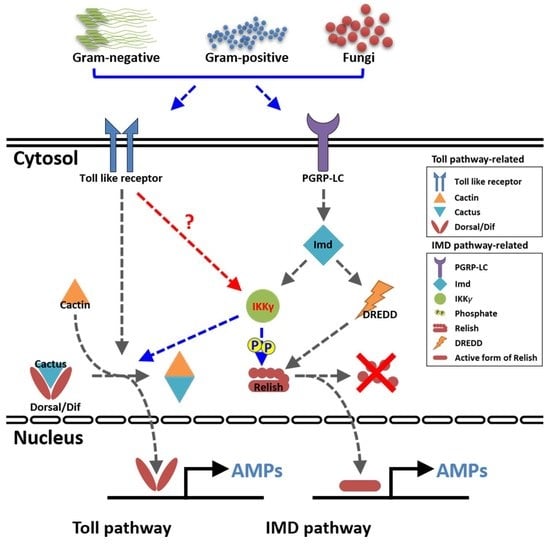
:1. Introduction
2. Results
2.1. Gene Organization and cDNA Structure of TmIKKγ
2.2. Domain Architecture and Phylogenetics of TmIKKγ
2.3. Developmental and Tissue-Specific Expression Patterns of TmIKKγ mRNA
2.4. Temporal Expression of TmIKKγ mRNA after Microbial Infection
2.5. TmIKKγ RNAi and Larval Mortality Assay
2.6. Effects of TmIKKγ RNAi on the Expression of AMPs
2.7. Effects of TmIKKγ RNAi on NF-κB Genes
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Preparation of Microorganisms
4.3. Identification and in Silico Analysis of TmIKKγ
4.4. Developmental, Tissue-Specific, and Immune Induction of TmIKKγ mRNA
4.5. TmIKKγ RNAi
4.6. Mortality Assay
4.7. Effects of TmIKKγ RNAi on the Expression of AMP and NF-κB Genes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zambon, R.A.; Nandakumar, M.; Vakharia, V.N.; Wu, L.P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 7257–7262. [Google Scholar] [CrossRef] [Green Version]
- Vercammen, E.; Staal, J.; Beyaert, R. Sensing of Viral Infection and Activation of Innate Immunity by Toll-Like Receptor 3. Clin. Microbiol. Rev. 2008, 21, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Kotwal, G.J.; Hatch, S.; Marshall, W.L. Viral Infection: An Evolving Insight into the Signal Transduction Pathways Responsible for the Innate Immune Response. Adv. Virol. 2012, 2012, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshiumi, H.; Matsuo, A.; Matsumoto, M.; Seya, T. Pan-Vertebrate Toll-Like Receptors During Evolution. Curr. Genom. 2008, 9, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Roy, D.; Patnaik, E.; Nongthomba, U. Muscles provide protection during microbial infection by activating innate immune response pathways in Drosophila and zebrafish. Dis. Model. Mech. 2016, 9, 697–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loures, F.V.; Pina, A.; Felonato, M.; Araújo, E.F.; Leite, K.M.; Calich, V.L.G. Toll-Like Receptor 4 Signaling Leads to Severe Fungal Infection Associated with Enhanced Proinflammatory Immunity and Impaired Expansion of Regulatory T Cells. Infect. Immun. 2009, 78, 1078–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venugopal, P.G.; Nutman, T.B.; Semnani, R.T. Activation and regulation of Toll-Like Receptors (TLRs) by helminth parasites. Immunol. Res. 2008, 43, 252–263. [Google Scholar] [CrossRef] [Green Version]
- Fouzder, C.; Mukhuty, A.; Das, S.; Chattopadhyay, D. TLR Signaling on Protozoan and Helminthic Parasite Infection. In Toll-Like Receptors; IntechOpen: London, UK, 2020. [Google Scholar]
- Leopizzi, M.; Cocchiola, R.; Milanetti, E.; Raimondo, D.; Politi, L.; Giordano, C.; Scandurra, R.; d’Abusco, A.S. IKKα inibition by a glucosamine derivative enhances maspin expression in osteosarcoma cell line. Chem. Biol. Interact. 2017, 262, 19–28. [Google Scholar] [CrossRef]
- Cao, C.C.; Li, L.P.; Chen, W.; Zhu, Y.F.; Qi, Y.C.; Wang, X.D.; Wan, X.; Chen, X. Deficiency of IKKε inhibits inflammation and induces cardiac protection in high-fat diet-induced obesity in mice. Int. J. Mol. Med. 2014, 34, 244–252. [Google Scholar] [CrossRef] [Green Version]
- De Falco, F.; Di Giovanni, C.; Cerchia, C.; De Stefano, D.; Capuozzo, A.; Irace, C.; Iuvone, T.; Santamaria, R.; Carnuccio, R.; Lavecchia, A. Novel non-peptide small molecules preventing IKKβ/NEMO association inhibit NF-κB activation in LPS-stimulated J774 macrophages. Biochem. Pharmacol. 2016, 104, 83–94. [Google Scholar] [CrossRef]
- Jiang, Y.; Tian, M.; Lin, W.; Wang, X.; Wang, X. Protein Kinase Serine/Threonine Kinase 24 Positively Regulates Interleukin 17-Induced Inflammation by Promoting IKK Complex Activation. Front. Immunol. 2018, 9, 921. [Google Scholar] [CrossRef]
- Mettang, M.; Reichel, S.N.; Lattke, M.; Palmer, A.; Abaei, A.; Rasche, V.; Huber-Lang, M.; Baumann, B.; Wirth, T. IKK2/NF-κB signaling protects neurons after traumatic brain injury. FASEB J. 2018, 32, 1916–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanji, T.; Hu, X.; Weber, A.N.R.; Ip, Y.T. Toll and IMD Pathways Synergistically Activate an Innate Immune Response in Drosophila melanogaster. Mol. Cell. Boil. 2007, 27, 4578–4588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoi, K.; Koyama, H.; Ito, W.; Minakuchi, C.; Tanaka, T.; Miura, K. Involvement of NF-κB transcription factors in antimicrobial peptide gene induction in the red flour beetle, Tribolium castaneum. Dev. Comp. Immunol. 2012, 38, 342–351. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef] [Green Version]
- Costechareyre, D.; Capo, F.; Fabre, A.; Chaduli, D.; Kellenberger, C.; Roussel, A.; Charroux, B.; Royet, J. Tissue-Specific Regulation of Drosophila NF-x03BA;B Pathway Activation by Peptidoglycan Recognition Protein SC. J. Innate Immun. 2015, 8, 67–80. [Google Scholar] [CrossRef]
- Iatsenko, I.; Kondo, S.; Mengin-Lecreulx, D.; Lemaitre, B. PGRP-SD, an Extracellular Pattern-Recognition Receptor, Enhances Peptidoglycan-Mediated Activation of the Drosophila Imd Pathway. Immunity 2016, 45, 1013–1023. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Shen, L.; Sun, L.; Xu, J.; Jin, P.; Chen, L.; Ma, F. Small RNA-Seq analysis reveals microRNA-regulation of the Imd pathway during Escherichia coli infection in Drosophila. Dev. Comp. Immunol. 2017, 70, 80–87. [Google Scholar] [CrossRef]
- Cheng, T.; Zhang, Y.L.; Liu, C.; Xu, P.Z.; Gao, Z.H.; Xia, Q.; Xiang, Z.H. Identification and analysis of Toll-related genes in the domesticated silkworm, Bombyx mori. Dev. Comp. Immunol. 2008, 32, 464–475. [Google Scholar] [CrossRef]
- Liu, W.; Liu, J.; Lu, Y.; Gong, Y.; Zhu, M.; Chen, F.; Liang, Z.; Zhu, L.; Kuang, S.; Hu, X.; et al. Immune signaling pathways activated in response to different pathogenic micro-organisms in Bombyx mori. Mol. Immunol. 2015, 65, 391–397. [Google Scholar] [CrossRef]
- Roh, K.B.; Kim, C.H.; Lee, H.; Kwon, H.M.; Park, J.W.; Ryu, J.H.; Kurokawa, K.; Ha, N.C.; Lee, W.J.; Lemaitre, B.; et al. Proteolytic Cascade for the Activation of the Insect Toll Pathway Induced by the Fungal Cell Wall Component. J. Boil. Chem. 2009, 284, 19474–19481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudzic, J.P.; Hanson, M.A.; Iatsenko, I.; Kondo, S.; Lemaitre, B. More Than Black or White: Melanization and Toll Share Regulatory Serine Proteases in Drosophila. Cell Rep. 2019, 27, 1050–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, J.H.; Ha, E.M.; Lee, W.J. Innate immunity and gut–microbe mutualism in Drosophila. Dev. Comp. Immunol. 2010, 34, 369–376. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.H.; Rämet, M. The Drosophila Toll Signaling Pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, U.N.; Yamamoto, Y.; Prajapati, S.; Gaynor, R.B. Nuclear role of IκB Kinase-γ/NF-κB Essential Modulator (IKKγ/NEMO) in NF-κB-dependent gene expression. J. Biol. Chem. 2004, 279, 3509–3515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Toth, Z.; Wong, L.Y.; Brulois, K.; Nguyen, J.; Lee, J.Y.; Zandi, E.; Jung, J.U. Novel phosphorylations of IKKγ/NEMO. MBio 2012, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudolph, D.; Yeh, W.C.; Wakeham, A.; Rudolph, B.; Nallainathan, D.; Potter, J.; Elia, A.J.; Mak, T.W. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKγ -deficient mice. Genes Dev. 2000, 14, 854–862. [Google Scholar] [PubMed]
- Rutschmann, S.; Jung, A.C.; Zhou, R.; Silverman, N.; Hoffmann, J.A.; Ferrandon, D. Role of Drosophila IKKγ in a Toll-independent antibacterial immune response. Nat. Immunol. 2000, 1, 342–347. [Google Scholar] [CrossRef]
- Kim, C.H.; Kim, S.J.; Kan, H.; Kwon, H.M.; Roh, K.B.; Jiang, R.; Yang, Y.; Park, J.W.; Lee, H.H.; Ha, N.C.; et al. A Three-step Proteolytic Cascade Mediates the Activation of the Peptidoglycan-induced Toll Pathway in an Insect. J. Boil. Chem. 2008, 283, 7599–7607. [Google Scholar] [CrossRef] [Green Version]
- Patnaik, B.B.; Patnaik, H.H.; Seo, G.W.; Jo, Y.H.; Lee, Y.S.; Lee, B.L.; Han, Y.S. Gene structure, cDNA characterization and RNAi-based functional analysis of a myeloid differentiation factor 88 homolog in Tenebrio molitor larvae exposed to Staphylococcus aureus infection. Dev. Comp. Immunol. 2014, 46, 208–221. [Google Scholar] [CrossRef]
- Jo, Y.H.; Kim, Y.J.; Park, K.B.; Seong, J.H.; Kim, S.G.; Park, S.; Noh, M.Y.; Lee, Y.S.; Han, Y.S. TmCactin plays an important role in Gram-negative and -positive bacterial infection by regulating expression of 7 AMP genes in Tenebrio molitor. Sci. Rep. 2017, 7, 46459. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Jo, Y.H.; Park, K.B.; Ko, H.J.; Edosa, T.T.; Lee, Y.S.; Han, Y. TmDorX2 positively regulates antimicrobial peptides in Tenebrio molitor gut, fat body, and hemocytes in response to bacterial and fungal infection. Sci. Rep. 2019, 9, 16878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Jo, Y.H.; Park, K.B.; Ko, H.J.; Kim, C.E.; Bae, Y.M.; Kim, B.; Jun, S.A.; Bang, I.S.; Lee, Y.S.; et al. TmToll-7 Plays a Crucial Role in Innate Immune Responses Against Gram-Negative Bacteria by Regulating 5 AMP Genes in Tenebrio molitor. Front. Immunol. 2019, 10, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, Y.H.; Patnaik, B.B.; Hwang, J.; Park, K.B.; Ko, H.J.; Kim, C.E.; Bae, Y.M.; Jung, W.J.; Lee, Y.S.; Han, Y. Regulation of the expression of nine antimicrobial peptide genes by TmIMD confers resistance against Gram-negative bacteria. Sci. Rep. 2019, 9, 10138. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Jo, Y.; Edosa, T.; Han, Y. Two Roles for the Tenebrio molitor Relish in the Regulation of Antimicrobial Peptides and Autophagy-Related Genes in Response to Listeria monocytogenes. Insects 2020, 11, 188. [Google Scholar] [CrossRef] [Green Version]
- Keshavarz, M.; Jo, Y.H.; Patnaik, B.B.; Park, K.B.; Ko, H.J.; Kim, C.E.; Edosa, T.T.; Lee, Y.S.; Han, Y.S. Tmrelish is required for regulating the antimicrobial responses to Escherichia coli and Staphylococcus aureus in Tenebrio molitor. Sci. Rep. 2020, 10, 4258. [Google Scholar] [CrossRef] [Green Version]
- Ertürk-Hasdemir, D.; Broemer, M.; Leulier, F.; Lane, W.S.; Paquette, N.; Hwang, D.; Kim, C.-H.; Stöven, S.; Meier, P.; Silverman, N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl. Acad. Sci. USA 2009, 106, 9779–9784. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.H.; Gu, Z.H.; Wan, D.H.; Liu, B.D.; Huang, X.D.; Weng, S.P.; Yu, X.Q.; He, J.G. The shrimp IKK–NF-κB signaling pathway regulates antimicrobial peptide expression and may be subverted by white spot syndrome virus to facilitate viral gene expression. Cell Mol. Immunol. 2013, 10, 423–436. [Google Scholar] [CrossRef]
- Baldwin, A.S. The NF-κB and IκB proteins: New discoveries and insights. Annu. Rev. Immunol. 1996, 14, 649–683. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Massa, P.E.; Hanidu, A.; Peet, G.W.; Aro, P.; Savitt, A.; Mische, S.; Li, J.; Marcu, K.B. IKKα, IKKβ, and NEMO/IKKγ are each required for the NF-κB-mediated inflammatory response program. J. Biol. Chem. 2002, 277, 45129–45140. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.Q.; Kovalenko, A.; Cantarella, G.; Wallach, D. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKγ) upon receptor stimulation. Immunity 2000, 12, 301–311. [Google Scholar] [CrossRef] [Green Version]
- Tusco, R.; Jacomin, A.-C.; Jain, A.; Penman, B.; Larsen, K.B.; Johansen, T.; Nezis, I.P. Kenny mediates selective autophagic degradation of the IKK complex to control innate immune responses. Nat. Commun. 2017, 8, 1264. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yu, M.; Chen, H.; Zeng, M.; Sun, Y.; Huang, Q. Identification and functional characterization of nemo in Crassostrea gigas reveals its crucial role in the NF-κB activation. Fish Shellfish Immunol. 2018, 80, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wu, C.J.; Zhao, Y.; Ashwell, J.D. Optineurin negatively regulates TNFα- Induced NF-κB activation by competing with nemo for ubiquitinated rip. Curr. Biol. 2007, 17, 1438–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, E.D.; Wang, C.Y.; Xiong, Y.; Guan, K.L. A role for NF-κB Essential Modifier/IκB Kinase-γ (NEMO/IKKγ) IκB Kinase Complex by Tumor Necrosis Factor-α. J. Biol. Chem. 2003, 278, 37297–37305. [Google Scholar] [CrossRef] [Green Version]
- Kalvari, I.; Tsompanis, S.; Mulakkal, N.C.; Osgood, R.; Johansen, T.; Nezis, I.P.; Promponas, V.J. Ilir: A web resource for prediction of atg8-family interacting proteins. Autophagy 2014, 10, 913–925. [Google Scholar] [CrossRef] [Green Version]
- Jacomin, A.-C.; Samavedam, S.; Promponas, V.J.; Nezis, I.P. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy 2016, 12, 1945–1953. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, S.; Aggarwal, K.; Paquette, N.; Silverman, N. NF-κB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr. Top. Microbiol. Immunol. 2011, 349, 25–60. [Google Scholar] [PubMed] [Green Version]
- Kleino, A.; Silverman, N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2013, 42, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Park, J.W.; Kwon, H.M.; Hwang, H.O.; Jang, I.H.; Masuda, A.; Kurokawa, K.; Nakayama, H.; Lee, W.J.; Dohmae, N.; et al. Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan in insects. J. Biol. Chem. 2010, 285, 32937–32945. [Google Scholar] [CrossRef] [Green Version]
- Hedengren-Olcott, M.; Olcott, M.C.; Mooney, D.T.; Ekengren, S.; Geller, B.L.; Taylor, B.J. Differential activation of the NF-κB-like factors relish and dif in Drosophila melanogaster by fungi and gram-positive bacteria. J. Biol. Chem. 2004, 279, 21121–21127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoi, K.; Koyama, H.; Minakuchi, C.; Tanaka, T.; Miura, K. Antimicrobial peptide gene induction, involvement of Toll and IMD pathways and defense against bacteria in the red flour beetle, Tribolium castaneum. Results Immunol. 2012, 2, 72–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, M.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.; McWilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; López, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]









| Name | Primer Sequences |
|---|---|
| TmIKKγ_cloning_Fw | 5′-TTGTTGTTCTGGGACCTTCC-3′ |
| TmIKKγ_cloning_Rv | 5′-GAGGGTGGTTGTTTGCAGTT-3′ |
| TmIKKγ_qPCR_Fw | 5′-TTGCGCAATTCCAAGGTGAC-3′ |
| TmIKKγ_qPCR_Rv | 5′-AGCCCCACAATTTCGGTTTG-3′ |
| dsTmIKKγ_Fw | 5′-TAATACGACTCACTATAGGGT |
| ATTTCCAGAGGCACCAACAG-3′ | |
| dsTmIKKγ_Rv | 5′-TAATACGACTCACTATAGGGT |
| AACTTGCTCTCTGCCAGCTC-3′ | |
| dsEGFP_Fw | 5′-TAATACGACTCACTATAGGG |
| TACGTAAACGGCCACAAGTTC-3′ | |
| dsEGFP_Rv | 5′-TAATACGACTCACTATAGGG |
| TTGCTCAGGTAGTGTTGTCG-3′ | |
| TmTenecin1_Fw | 5′-CAGCTGAAGAAATCGAACAAGG-3′ |
| TmTenecin1_Rv | 5′-CAGACCCTCTTTCCGTTACAGT-3′ |
| TmTenecin2_Fw | 5′-CAGCAAAACGGAGGATGGTC-3′ |
| TmTenecin2_Rv | 5′-CGTTGAAATCGTGATCTTGTCC-3′ |
| TmTenecin3_Fw | 5′-GATTTGCTTGATTCTGGTGGTC-3′ |
| TmTenecin3_Rv | 5′-CTGATGGCCTCCTAAATGTCC-3′ |
| TmTenecin4_Fw | 5′-GGACATTGAAGATCCAGGAAAG-3′ |
| TmTenecin4_Rv | 5′-CGGTGTTCCTTATGTAGAGCTG-3′ |
| TmDefensin1_Fw | 5′-AAATCGAACAAGGCCAACAC-3′ |
| TmDefencin1_Rv | 5′-GCAAATGCAGACCCTCTTTC-3′ |
| TmDefensin2_Fw | 5′-GGGATGCCTCATGAAGATGTAG-3′ |
| TmDefensin2_Rv | 5′-CCAATGCAAACACATTCGTC-3′ |
| TmColeoptericin1_Fw | 5′-GGACAGAATGGTGGATGGTC-3′ |
| TmColeoptericin1_Rv | 5′-CTCCAACATTCCAGGTAGGC-3′ |
| TmColeoptericin2_Fw | 5′-GGACGGTTCTGATCTTCTTGAT-3′ |
| TmColeoptericin2_Rv | 5′-CAGCTGTTTGTTTGTTCTCGTC-3′ |
| TmAttacin1a_Fw | 5′-GAAACGAAATGGAAGGTGGA-3′ |
| TmAttacin1a_Rv | 5′-TGCTTCGGCAGACAATACAG-3′ |
| TmAttacin1b_Fw | 5′-GAGCTGTGAATGCAGGACAA-3′ |
| TmAttacin1b_Rv | 5′-CCCTCTGATGAAACCTCCAA-3′ |
| TmAttacin2_Fw | 5′-AACTGGGATATTCGCACGTC-3′ |
| TmAttacin2_Rv | 5′-CCCTCCGAAATGTCTGTTGT-3′ |
| TmCecropin2_Fw | 5′-TACTAGCAGCGCCAAAACCT-3′ |
| TmCecropin2_Rv | 5′-CTGGAACATTAGGCGGAGAA-3′ |
| TmThaumatin-like protein1_Fw | 5′-CTCAAAGGACACGCAGGACT-3′ |
| TmThaumatin-like protein1_Rv | 5′-ACTTTGAGCTTCTCGGGACA-3′ |
| TmThaumatin-like protein2_Fw | 5′-CCGTCTGGCTAGGAGTTCTG-3′ |
| TmThaumatin-like protein2_Rv | 5′-ACTCCTCCAGCTCCGTTACA-3′ |
| TmDorX1_qPCR_Fw | 5′-AGCGTTGAGGTTTCGGTATG-3′ |
| TmDorX1_qPCR_Rv | 5′-TCTTTGGTGACGCAAGACAC-3′ |
| TmDorX2_qPCR_Fw | 5′-ACACCCCCGAAATCACAAAC-3′ |
| TmDorX2_qPCR_Rv | 5′-TTTCAGAGCGCCAGGTTTTG-3′ |
| TmRelish_qPCR_Fw | 5′-AGCGTCAAGTTGGAGCAGAT-3′ |
| TmRelish_qPCR_Rv | 5′-GTCCGGACCTCAAGTGT-3′ |
| TmL27a_qPCR_Fw | 5′-TCATCCTGAAGGCAAAGCTCCAGT-3′ |
| TmL27a_qPCR_Rv | 5′-AGGTTGGTTAGGCAGGCACCTTTA-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, H.J.; Jo, Y.H.; Patnaik, B.B.; Park, K.B.; Kim, C.E.; Keshavarz, M.; Jang, H.A.; Lee, Y.S.; Han, Y.S. IKKγ/NEMO Is Required to Confer Antimicrobial Innate Immune Responses in the Yellow Mealworm, Tenebrio Molitor. Int. J. Mol. Sci. 2020, 21, 6734. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186734
Ko HJ, Jo YH, Patnaik BB, Park KB, Kim CE, Keshavarz M, Jang HA, Lee YS, Han YS. IKKγ/NEMO Is Required to Confer Antimicrobial Innate Immune Responses in the Yellow Mealworm, Tenebrio Molitor. International Journal of Molecular Sciences. 2020; 21(18):6734. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186734
Chicago/Turabian StyleKo, Hye Jin, Yong Hun Jo, Bharat Bhusan Patnaik, Ki Beom Park, Chang Eun Kim, Maryam Keshavarz, Ho Am Jang, Yong Seok Lee, and Yeon Soo Han. 2020. "IKKγ/NEMO Is Required to Confer Antimicrobial Innate Immune Responses in the Yellow Mealworm, Tenebrio Molitor" International Journal of Molecular Sciences 21, no. 18: 6734. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186734