Temperature Regulation of Primary and Secondary Seed Dormancy in Rosa canina L.: Findings from Proteomic Analysis
Abstract
:1. Introduction
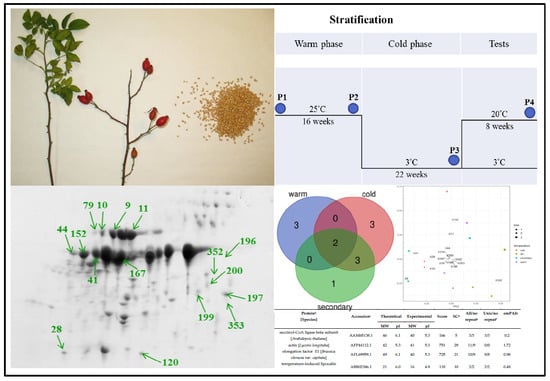
2. Results and Discussion
3. Materials and Methods
3.1. Plant Material
3.2. Seed Germination
3.3. Proteome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kendall, S.; Penfield, S. Maternal and zygotic temperature signalling in the control of seed dormancy and germination. Seed Sci. Res. 2012, 22, S23–S29. [Google Scholar] [CrossRef]
- Hilhorst, H.W.M. The regulation of secondary dormancy. The membrane hypothesis revisite. Seed Sci. Res. 1998, 8, 77–90. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. N. Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Huang, Z.; Clay, H.A.; Mead, A.; Finch-Savage, W.E. Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant. J. 2013, 74, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Murphey, M.; Kovach, K.; Elnacash, T.; He, H.; Bentsink, L.; Donohue, K. DOG1-imposed dormancy mediates germination responses to temperature cues. Environ. Exp. Bot. 2015, 112, 33–43. [Google Scholar] [CrossRef]
- Soltani, E.; Baskin, J.M.; Baskin, C.C. A review of the relationship between primary and secondary dormancy, with reference to the volunteer crop weed oilseed rape (Brassica napus). Weed Res. 2019, 59, 5–14. [Google Scholar] [CrossRef] [Green Version]
- Footitt, S.; Douterelo-Soler, I.; Clay, H.; Finch-Savage, W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 20236–20241. [Google Scholar] [CrossRef] [Green Version]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. N. Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [Green Version]
- Staszak, A.M.; Rewers, M.; Sliwinska, E.; Klupczynska, E.A.; Pawlowski, T. DNA synthesis pattern, proteome, and ABA and GA signalling in developing seeds of Norway maple (Acer platanoides). Funct. Plant Biol. 2019, 46, 152–164. [Google Scholar] [CrossRef]
- Basbouss-Serhal, I.; Leymarie, J.; Bailly, C. Fluctuation of Arabidopsis seed dormancy with relative humidity and temperature during dry storage. J. Exp. Bot. 2016, 67, 119–130. [Google Scholar] [CrossRef]
- Chang, G.; Wang, C.; Kong, X.-X.; Chen, Q.; Yang, Y.; Hu, X. AFP2 as the novel regulator breaks high-temperature-induced seeds secondary dormancy through ABI5 and SOM in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 501, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.S.; Saleh, Y.; Gazzarrini, S. Inhibition of FUSCA3 degradation at high temperature is dependent on ABA signaling and is regulated by the ABA/GA ratio. Plant Signal. Behav. 2016, 11, e1247137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibarra, S.E.; Tognacca, R.S.; Dave, A.; Graham, I.A.; Sánchez, R.A.; Botto, J.F. Molecular mechanisms underlying the entrance in secondary dormancy of Arabidopsis seeds. Plant Cell Environ. 2015, 39, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kępczyński, J.; Bihun, M.; Kępczyñska, E. Implication of ethylene in the release of secondary dormancy in Amaranthus caudatus L. seeds by gibberellins or cytokinin. Plant Growth Regul. 2006, 48, 119–126. [Google Scholar] [CrossRef]
- Kępczyński, J.; Cembrowska-Lech, D.; Sznigir, P. Interplay between nitric oxide, ethylene, and gibberellic acid regulating the release of Amaranthus retroflexus seed dormancy. Acta Physiol. Plant 2017, 39, 254. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, J.; Liu, Y.; Zhao, J.; Fu, J.; Ren, X.; Wang, G.; Wang, J. Exogenous auxin regulates multi-metabolic network and embryo development, controlling seed secondary dormancy and germination in Nicotiana tabacum L. BMC Plant Biol. 2016, 16, 41. [Google Scholar] [CrossRef] [Green Version]
- Footitt, S.; Ölçer-Footitt, H.; Hambidge, A.J.; Finch-Savage, W.E. A laboratory simulation of Arabidopsis seed dormancy cycling provides new insight into its regulation by clock genes and the dormancy-related genes DOG1, MFT, CIPK23 and PHYA. Plant Cell Environ. 2017, 40, 1474–1486. [Google Scholar] [CrossRef] [Green Version]
- Née, G.; Obeng-Hinneh, E.; Sarvari, P.; Nakabayashi, K.; Soppe, W.J. Secondary dormancy in Brassica napus is correlated with enhanced BnaDOG1 transcript levels. Seed Sci. Res. 2015, 25, 221–229. [Google Scholar] [CrossRef]
- Chiang, G.C.K.; Barua, D.; Dittmar, E.; Kramer, E.M.; De Casas, R.R.; Donohue, K. Pleiotropy in the wild: The dormancy gene DOG1 exerts cascading control on life cycles. Evolution 2013, 67, 883–893. [Google Scholar] [CrossRef]
- Suszka, B.; Bujarska-Borkowska, B. After-ripening, germination of seeds and seedling emergence of Rosa canina L. “Schmids Ideal” in relation to other rootstock selection of this species. Arboretum Kórnickie 1989, 34, 113–134. [Google Scholar]
- Suszka, B.; Bujarska-Borkowska, B. Seed after-ripening, germination and seedling emergence of Rosa canina L. and some of its rootstock selection. Arboretum Kórnickie 1987, 32, 231–296. [Google Scholar]
- Hilhorst, H.W. Definitions and hypotheses of seed dormancy. In Seed Development, Dormancy and Germination; Bradford, K.J., Nonogaki, H., Eds.; Wiley-Blackwell: Oxford, UK, 2007; pp. 50–71. [Google Scholar]
- Edwards, B.; Burghardt, L.T.; Kovach, K.E.; Donohue, K. Canalization of seasonal phenology in the presence of developmental variation: Seed dormancy cycling in an annual weed. Integr. Comp. Biol. 2017, 57, 1021–1039. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, F.; Chu, J.; Yi, X.; Fan, W.; Tang, T.; Chen, G.; Guo, Q.; Zhao, X. A transcriptome analysis reveals a role for the indole GLS-linked auxin biosynthesis in secondary dormancy in rapeseed (Brassica napus L.). BMC Plant Biol. 2019, 19, 264. [Google Scholar] [CrossRef] [PubMed]
- Fei, H.; Tsang, E.; Cutler, A.J. Gene expression during seed maturation in Brassica napus in relation to the induction of secondary dormancy. Genomics 2007, 89, 419–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Zhao, X.; Zhang, L.; Tang, T.; Lu, C.; Chen, G.; Wang, X.; Bu, C.; Zhao, X. RNA-seq profiling the transcriptome of secondary seed dormancy in canola (Brassica napus L.). Chin. Sci. Bull. 2014, 59, 4341–4351. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified Protein Abundance Index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Staszak, A.M.; Pawlowski, T.A. Forest tree research in post genomic era. Introduction to systems biology of broadleaves. Dendrobiology 2012, 68, 113–123. [Google Scholar]
- Pawlowski, T.; Bergervoet, J.; Bino, R.; Groot, S.P.C. Cell cycle activity and β-tubulin accumulation during dormancy breaking of Acer platanoides L. seeds. Biol. Plant. 2004, 48, 211–218. [Google Scholar] [CrossRef]
- Bentsink, L.; Koornneef, M. Seed dormancy and germination. Arab. Book 2008, 6, e0119. [Google Scholar] [CrossRef] [Green Version]
- Née, G.; Xiang, Y.; Soppe, W.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant. Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.L.; De Diego, J.G.; Rodríguez, F.D.; Cervantes, E. Mitochondrial structures during seed germination and early seedling development in Arabidopsis thaliana. Biologia 2015, 70, 1019–1025. [Google Scholar] [CrossRef]
- Czarna, M.; Kolodziejczak, M.; Janska, H. Mitochondrial proteome studies in seeds during germination. Proteomes 2016, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Shotwell, M.A.; Afonso, C.; Davies, E.; Chesnut, R.S.; Larkins, B.A. Molecular characterization of oat seed globulins. Plant Physiol. 1988, 87, 698–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prewein, C.; Endemann, M.; Reinöhl, V.; Salaj, J.; Sunderlikova, V.; Wilhelm, E. Physiological and morphological characteristics during development of pedunculate oak (Quercus robur L.) zygotic embryos. Trees 2006, 20, 53–60. [Google Scholar] [CrossRef]
- Gruis, D.F.; Selinger, D.A.; Curran, J.M.; Jung, R. Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 2002, 14, 2863–2882. [Google Scholar] [CrossRef]
- Krasuska, U.; Ciacka, K.; Orzechowski, S.; Fettke, J.; Bogatek, R.; Gniazdowska, A. Modification of the endogenous NO level influences apple embryos dormancy by alterations of nitrated and biotinylated protein patterns. Planta 2016, 244, 877–891. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Gilliland, L.U.; McKinney, E.C.; Meagher, R.B. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 2001, 13, 1541–1554. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, K.; Le Signor, C.; Vandekerckhove, J.; Thompson, R.D.; Burstin, J. Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 2003, 133, 664–682. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.J.; Hu, X.F.; Ai, X.R.; Yao, L.; Deng, S.M.; Pu, X.; Song, S.Q. Dormancy release of Cotinus coggygria seeds under a pre-cold moist stratification: An endogenous abscisic acid/gibberellic acid and comparative proteomic analysis. N. For. 2016, 47, 105–118. [Google Scholar] [CrossRef]
- Díaz-Camino, C.; Conde, R.; Ovsenek, N.; Villanueva, M.A. Actin expression is induced and three isoforms are differentially expressed during germination in Zea mays. J. Exp. Bot. 2005, 56, 557–565. [Google Scholar] [CrossRef] [Green Version]
- De Farias, E.T.; Da Silva, E.A.A.; Toorop, P.E.; Bewley, J.D.; Hilhorst, H.W.M. Expression studies in the embryo and in the micropylar endosperm of germinating coffee (Coffea arabica cv. Rubi) seeds. Plant Growth Regul. 2015, 75, 575–581. [Google Scholar] [CrossRef] [Green Version]
- Gilliland, L.U.; Pawloski, L.C.; Kandasamy, M.K.; Meagher, R.B. Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 2003, 33, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Park, M.R.; Wang, Y.-H.; Hasenstein, K.H. Profiling gene expression in germinating Brassica roots. Plant. Mol. Biol. Rep. 2014, 32, 541–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.-Q.; Song, B.-Y.; Deng, Z.-J.; Wang, Y.; Liu, S.-J.; Møller, I.M.; Song, S.-Q. Proteomic analysis of lettuce seed germination and thermoinhibition by sampling of individual seeds at germination and removal of storage proteins by polyethylene glycol fractionation. Plant Physiol. 2015, 167, 1332–1350. [Google Scholar] [CrossRef]
- Haferkamp, I.; Fernie, A.R.; Neuhaus, H.E. Adenine nucleotide transport in plants: Much more than a mitochondrial issue. Trends Plant Sci. 2011, 16, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca-Pereira, P.; Neri-Silva, R.; Cavalcanti, J.H.F.; Brito, D.S.; Weber, A.P.; Araújo, W.L.; Nunes-Nesi, A. Data-mining bioinformatics: Connecting adenylate transport and metabolic responses to stress. Trends Plant Sci. 2018, 23, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Plocharski, B.; Haferkamp, I.; Leroch, M.; Ewald, R.; Bauwe, H.; Riemer, J.; Herrmann, J.M.; Neuhaus, H.E. From endoplasmic reticulum to mitochondria: Absence of the Arabidopsis ATP antiporter endoplasmic reticulum adenylate transporter1 perturbs photorespiration. Plant Cell 2013, 25, 2647–2660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroch, M.; Neuhaus, H.E.; Kirchberger, S.; Zimmermann, S.; Melzer, M.; Gerhold, J.; Tjaden, J. Identification of a Novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. Plant Cell 2008, 20, 438–451. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.-F.; Chen, P.-Y.; Nguyen, Q.T.T.; Huang, L.-Y.; Zeng, G.-R.; Huang, T.-L.; Lin, C.-Y.; Huang, H. Transcriptome profiling of genes and pathways associated with arsenic toxicity and tolerance in Arabidopsis. BMC Plant Biol. 2014, 14, 94. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, F.; Pierri, C.L.; De Grassi, A.; Nunes-Nesi, A.; Fernie, A.R. Evolution, structure and function of mitochondrial carriers: A review with new insights. Plant J. 2011, 66, 161–181. [Google Scholar] [CrossRef]
- Lo, Y.-S.; Cheng, N.; Hsiao, L.-J.; Annamalai, A.; Jauh, G.-Y.; Wen, T.-N.; Dai, H.; Chiang, K.-S. Actin in mung bean mitochondria and implications for its function. Plant Cell 2011, 23, 3727–3744. [Google Scholar] [CrossRef] [Green Version]
- Senior, A.E.; Nadanaciva, S.; Weber, J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta Bioenerg. 2002, 1553, 188–211. [Google Scholar] [CrossRef] [Green Version]
- Zaynab, M.; Pan, D.; Noman, A.; Fatima, M.; Abbas, S.; Umair, M.; Sharif, Y.; Chen, S.-P.; Chen, W. Transcriptome approach to address low seed germination in Cyclobalanopsis gilva to save forest ecology. Biochem. Syst. Ecol. 2018, 81, 62–69. [Google Scholar] [CrossRef]
- He, M.; Zhu, C.; Dong, K.; Zhang, T.; Cheng, Z.; Li, J.; Yan, Y. Comparative proteome analysis of embryo and endosperm reveals central differential expression proteins involved in wheat seed germination. BMC Plant Biol. 2015, 15, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlowski, T. Proteomics of European beech (Fagus sylvatica L.) seed dormancy breaking: Influence of abscisic and gibberellic acids. Proteomics 2007, 7, 2246–2257. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, T. Proteome analysis of Norway maple (Acer platanoides L.) seeds dormancy breaking and germination: Influence of abscisic and gibberellic acids. BMC Plant Biol. 2009, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Pawlowski, T.; Staszak, A.M. Analysis of the embryo proteome of sycamore (Acer pseudoplatanus L.) seeds reveals a distinct class of proteins regulating dormancy release. J. Plant Physiol. 2016, 195, 9–22. [Google Scholar] [CrossRef]
- Wojtyla, Ł.; Kosmala, A.; Garnczarska, M. Lupine embryo axes under salinity stress. II. Mitochondrial proteome response. Acta Physiol. Plant 2013, 35, 2383–2392. [Google Scholar] [CrossRef] [Green Version]
- Yin, G.; Sun, H.; Xin, X.; Qin, G.; Liang, Z.; Jing, X. Mitochondrial damage in the soybean seed axis during imbibition at chilling temperatures. Plant Cell Physiol. 2009, 50, 1305–1318. [Google Scholar] [CrossRef] [Green Version]
- Pawlowski, T. Proteomic approach to analyze dormancy breaking of tree seeds. Plant. Mol. Biol. 2010, 73, 15–25. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, T.; Staszak, A.M.; Karolewski, P.; Giertych, M.J. Plant development reprogramming by cynipid gall wasp: Proteomic analysis. Acta Physiol. Plant 2017, 39, 114. [Google Scholar] [CrossRef]


| Spot a | Protein b [Species] | Accession c | Theoretical | Experimental | Score | SC d | All/No Repeat e | Unic/No Repeat f | emPAI g | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | pI | MW | pI | ||||||||
| 9 | succinyl-CoA ligase beta subunit [Arabidopsis thaliana] | AAM65138.1 | 46 | 6.1 | 40 | 5.3 | 166 | 5 | 3/3 | 3/3 | 0.2 |
| 10 | actin [Lycoris longituba] | AFP44112.1 | 42 | 5.3 | 41 | 5.3 | 751 | 29 | 11/9 | 0/0 | 1.72 |
| 11 | elongation factor E1 [Brassica oleracea var. capitata] | AFL69959.1 | 49 | 6.1 | 40 | 5.3 | 725 | 21 | 10/9 | 9/8 | 0.98 |
| 28 | temperature-induced lipocalin [Solanum tuberosum] | ABB02386.1 | 21 | 6.0 | 16 | 4.9 | 118 | 10 | 2/2 | 2/2 | 0.48 |
| 41 | legumin B-like [Fragaria vesca subsp. vesca] | XP_004294115.1 | 57 | 6.8 | 36 | 5.2 | 483 | 16 | 31/6 | 16/3 | 0.56 |
| 44 | adenosine kinase 2 [Glycine soja] | KHN02332.1 | 38 | 5.5 | 37 | 5 | 99 | 3 | 1/1 | 1/1 | 0.12 |
| 79 | actin-7 [Musa acuminata subsp. malaccensis] | XP_009383456.1 | 42 | 5.3 | 41 | 5.2 | 350 | 16 | 5/5 | 0/0 | 0.65 |
| 120 | cytosolic class I small heat-shock protein HSP17.5 [Rosa hybrid cultivar] | ABO84841.1 | 17 | 6.0 | 15 | 5.8 | 562 | 46 | 16/8 | 2/2 | 5.64 |
| 152 | mitochondrial ADP/ATP translocator [Chlamydomonas incerta] | ABA01103.1 | 34 | 9.7 | 37 | 5.0 | 357 | 15 | 5/5 | 1/1 | 0.86 |
| 167 | legumin B-like [F. vesca subsp. vesca] | XP_004294115.1 | 57 | 6.3 | 36 | 5.5 | 493 | 16 | 40/6 | 13/3 | 0.56 |
| 196 | 2-dehydro-3-deoxyphosphooctonate aldolase 1 [F. vesca subsp. vesca] | XP_004306551.1 | 32 | 6.6 | 32 | 6.1 | 590 | 33 | 9/8 | 9/9 | 2.24 |
| 197 | ATPase alpha subunit, partial (mitochondrion) [Chlorokybus atmophyticus] | ABI54626.1 | 38 | 9.3 | 24 | 7 | 131 | 5 | 2/2 | 2/2 | 0.12 |
| 199 | triosephosphate isomerase, cytosolic [Zea mays] | ACG24648.1 | 27 | 5.5 | 23 | 6.6 | 283 | 14 | 3/3 | 1/1 | 0.58 |
| 200 | triosephosphate isomerase, cytosolic [Z. mays] | ACG24648.1 | 27 | 5.5 | 25 | 6.8 | 374 | 19 | 4/4 | 2/2 | 0.85 |
| 352 | glyceraldehyde 3-phosphate dehydrogenase [R. multiflora] | AEQ75490.1 | 37 | 7.7 | 27 | 6.9 | 562 | 33 | 11/10 | 1/1 | 1.51 |
| 353 | oil body-associated protein 1A-like [R. chinensis] | XP_024167493.1 | 27 | 5.9 | 26 | 7.1 | 408 | 19 | 6/5 | 5/4 | 1.19 |
| Spot a | Protein b | Mean % Volume (± s.d.) c | |||
|---|---|---|---|---|---|
| Dry | Warm | Cold | Secondary | ||
| 28 | temperature-induced lipocalin | 0.09 ± 0.07 c | 0.22 ± 0.08 bc | 0.39 ± 0.21 ab | 0.46 ± 0.11 a |
| 41 | legumin B-like | 1.16 ± 0.21 b | 1.53 ± 0.30 b | 2.25 ± 0.49 a | 1.49 ± 0.38 b |
| 79 | actin-7 | 0.06 ± 0.01 b | 0.12 ± 0.02 ab | 0.20 ± 0.11 a | 0.15 ± 0.05 ab |
| 120 | cytosolic class I small heat-shock protein HSP17.5 | 0.09 ± 0.01 b | 0.08 ± 0.01 b | 0.12 ± 0.06 ab | 0.16 ± 0.03 a |
| 152 | mitochondrial ADP/ATP translocator | 0.63 ± 0.16 b | 0.85 ± 0.21 ab | 1.15 ± 0.24 a | 0.91 ± 0.20 ab |
| 167 | legumin B-like | 4.96 ± 0.60 a | 2.84 ± 0.82 ab | 2.38 ± 1.11 b | 2.23 ± 1.43 b |
| 196 | 2-dehydro-3-deoxyphosphooctonate aldolase 1 | 0.08 ± 0.04 a | 0.03 ± 0.04 ab | 0 b | 0 B |
| 197 | ATPase alpha subunit | 0 b | 0.11 ± 0.10 a | 0.07 ± 0.10 ab | 0.04 ± 0.06 ab |
| 199 | triosephosphate isomerase | 0 b | 0.05 ± 0.03 a | 0.001 ± 0.001 b | 0.02 ± 0.02 ab |
| 200 | triosephosphate isomerase | 0 b | 0.07 ± 0.02 a | 0.05 ± 0.07 ab | 0.06 ± 0.08 ab |
| 352 | glyceraldehyde 3-phosphate dehydrogenase | 0.05 ± 0.04 a | 0 b | 0 b | 0 B |
| 353 | oil body-associated protein 1A-like | 0.13 ± 0.21 a | 0 b | 0 b | 0 B |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pawłowski, T.A.; Bujarska-Borkowska, B.; Suszka, J.; Tylkowski, T.; Chmielarz, P.; Klupczyńska, E.A.; Staszak, A.M. Temperature Regulation of Primary and Secondary Seed Dormancy in Rosa canina L.: Findings from Proteomic Analysis. Int. J. Mol. Sci. 2020, 21, 7008. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197008
Pawłowski TA, Bujarska-Borkowska B, Suszka J, Tylkowski T, Chmielarz P, Klupczyńska EA, Staszak AM. Temperature Regulation of Primary and Secondary Seed Dormancy in Rosa canina L.: Findings from Proteomic Analysis. International Journal of Molecular Sciences. 2020; 21(19):7008. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197008
Chicago/Turabian StylePawłowski, Tomasz A., Barbara Bujarska-Borkowska, Jan Suszka, Tadeusz Tylkowski, Paweł Chmielarz, Ewelina A. Klupczyńska, and Aleksandra M. Staszak. 2020. "Temperature Regulation of Primary and Secondary Seed Dormancy in Rosa canina L.: Findings from Proteomic Analysis" International Journal of Molecular Sciences 21, no. 19: 7008. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197008