An Influence of Modification with Phosphoryl Guanidine Combined with a 2′-O-Methyl or 2′-Fluoro Group on the Small-Interfering-RNA Effect
Abstract
:1. Introduction
2. Results and Discussion
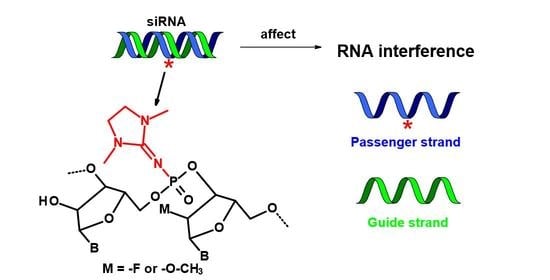
2.1. Design of PG-Modified siRNA
2.2. Duplex Thermostability
2.3. Stability of Modified siRNA in the Presence of RNase A
2.4. The Activity of Modifed siRNA In Vitro
3. Materials and Methods
3.1. Reagents
3.2. Oligonucleotide Synthesis
3.3. UV-Melting Experiments
3.4. 5′-End-32P-Labeling of Antisense siRNA Chains
3.5. 3’-End-Labeling of Antisense siRNA Chains with FITC
3.6. RNAse A Degradation Assays
3.7. Cell Culture and Transfection
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [Green Version]
- Almeida, R.; Allshire, R.C. RNA silencing and genome regulation. Trends Cell Biol. 2005, 15, 251–258. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial Rnase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Bernards, R. Exploring the uses of RNAi–Gene Knockdown and the Nobel Prize. N. Engl. J. Med. 2006, 355, 2391–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Agarwal, S.; Simon, A.R.; Goel, V.; Habtemariam, B.A.; Clausen, V.A.; Kim, J.B.; Robbie, G.J. Pharmacokinetics and Pharmacodynamics of the Small Interfering Ribonucleic Acid, Givosiran, in Patients With Acute Hepatic Porphyria. Clin. Pharmacol. Ther. 2020, 108, 63–72. [Google Scholar] [CrossRef]
- Shen, X.; Corey, D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018, 46, 1584–1600. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Sig. Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Stetsenko, D.A.; Kupryushkin, M.S.; Pyshnyi, D.V. Modified Oligonucleotides and Methods for Their Synthesis. International Patent No. WO2,016,028,187A1, 22 June 2014. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2016028187 (accessed on 7 September 2021).
- Lomzov, A.A.; Kupryushkin, M.S.; Shernyukov, A.V.; Nekrasov, M.D.; Dovydenko, I.S.; Stetsenko, D.A.; Pyshnyi, D.V. Diastereomers of a mono-substituted phosphoryl guanidine trideoxyribonucleotide: Isolation and properties. Biochem. Biophys. Res. Commun. 2019, 513, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Lomzov, A.A.; Kupryushkin, M.S.; Dyudeeva, E.S.; Pyshnyi, D.V. A Comparative Study of the Hybridization of Phosphoryl Guanidine Oligonucleotides with DNA and RNA. Russ. J. Bioorg. Chem. 2021, 47, 461–468. [Google Scholar] [CrossRef]
- Tschuch, C.; Schulz, A.; Pscherer, A.; Werft, W.; Benner, A.; Hotz-Wagenblatt, A.; Barrionuevo, L.S.; Lichter, P.; Mertens, D. Off-target effects of siRNA specific for GFP. BMC Mol. Biol. 2008, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Dyudeeva, E.S.; Pavlova, A.S.; Kupryushkin, M.S.; Pyshnyi, D.V.; Pyshnaya, I.A. Problems of the Synthesis of Oligonucleotide Derivatives in the Realization of the Anchimeric Effect. Russ. J. Bioorg. Chem. 2020, 47, 505–513. [Google Scholar] [CrossRef]
- Bennett, C.F.; Swayze, E.E. RNA Targeting Therapeutics: Molecular Mechanisms of Antisense Oligonucleotides as a Therapeutic Platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [Google Scholar] [CrossRef]
- Kawasaki, A.M.; Casper, M.D.; Freier, S.M.; Lesnik, E.A.; Zounes, M.C.; Cummins, L.L.; Gonzalez, C.; Cook, P.D. Uniformly modified 2’-deoxy-2’-fluoro-phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993, 36, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Volkov, A.A.; Kruglova, N.S.; Meschaninova, M.I.; Venyaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Selective Protection of Nuclease-Sensitive Sites in siRNA Prolongs Silencing Effect. Oligonucleotides 2009, 19, 191–202. [Google Scholar] [CrossRef]
- Lisowiec-Wąchnicka, J.; Bartyś, N.; Pasternak, A. A systematic study on the influence of thermodynamic asymmetry of 5′-ends of siRNA duplexes in relation to their silencing potency. Sci. Rep. 2019, 9, 2477. [Google Scholar] [CrossRef] [Green Version]
- Vert, J.-P.; Foveau, N.; Lajaunie, C.; Vandenbrouck, Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinform. 2006, 7, 520. [Google Scholar] [CrossRef] [Green Version]
- Raines, R.T. Ribonuclease A. Chem. Rev. 1998, 98, 1045–1066. [Google Scholar] [CrossRef]
- Iribe, H.; Miyamoto, K.; Takahashi, T.; Kobayashi, Y.; Leo, J.; Aida, M.; Ui-Tei, K. Chemical Modification of the siRNA Seed Region Suppresses Off-Target Effects by Steric Hindrance to Base-Pairing with Targets. ACS Omega 2017, 2, 2055–2064. [Google Scholar] [CrossRef] [Green Version]
- Martin-Pintado, N.; Deleavey, G.F.; Portella, G.; Campos-Olivas, R.; Orozco, M.; Damha, M.J.; González, C. Backbone FC–H⋅⋅⋅O Hydrogen Bonds in 2′F-Substituted Nucleic Acids. Angew. Chem. Int. Ed. Engl. 2013, 52, 12065–12068. [Google Scholar] [CrossRef] [PubMed]
- Bellon, L. Oligoribonucleotides with 2′-O-(tert-Butyldimethylsilyl) Groups. Curr. Protoc. Nucleic Acid Chem. 2000, 1, 3–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, N.S.; Chernikov, I.V.; Meschaninova, M.I.; Dovydenko, I.S.; Venyaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Carrier-free cellular uptake and the gene-silencing activity of the lipophilic siRNAs is strongly affected by the length of the linker between siRNA and lipophilic group. Nucleic Acids Res. 2012, 40, 2330–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dovydenko, I.; Heckel, A.-M.; Tonin, Y.; Gowher, A.; Venyaminova, A.; Tarassov, I.; Entelis, N. Mitochondrial Targeting of Recombinant RNA. In Mitochondrial Medicine. Methods in Molecular Biology; Weissig, V., Edeas, M., Eds.; Humana Press: New York, NY, USA, 2015; Volume 1265, pp. 209–225. [Google Scholar] [CrossRef]








| Row | Examined Duplex | Comparison Duplex | ΔTm, °C |
|---|---|---|---|
| 1 | As:S-OMe* | As:S-OMe | −1.4 |
| 2 | As:S-F* | As:S-F | −1.1 |
| 3 | As-OMe:S-OMe* | As-OMe:S-OMe | −1.4 |
| 4 | As-OMe:S-F* | As-OMe:S-F | −1.4 |
| 5 | As-OMe*:S-OMe* | As-OMe*:S-OMe | −1 |
| 6 | As-OMe*:S-F* | As-OMe*:S-F | −1.7 |
| 7 | As-F:S-OMe* | As-F:S-OMe | −0.4 |
| 8 | As-F:S-F* | As-F:S-F | −0.9 |
| 9 | As-F*:S-OMe* | As-F*:S-OMe | −0.6 |
| 10 | As-F*:S-F* | As-F*:S-F | −0.9 |
| 11 | As-OMe*:S | As-OMe:S | 0 |
| 12 | As-F*:S | As-F:S | −0.9 |
| 13 | As-OMe*:S-OMe | As-OMe:S-OMe | −0.3 |
| 14 | As-F*:S-OMe | As-F:S-OMe | −1.3 |
| 15 | As-OMe*:S-OMe* | As-OMe:S-OMe* | 0.1 |
| 16 | As-F*:S-OMe* | As-F:S-OMe* | −1.5 |
| 17 | As-OMe*:S-F | As-OMe:S-F | 0.4 |
| 18 | As-F*:S-F | As-F:S-F | −0.3 |
| 19 | As-OMe*:S-F* | As-OMe:S-F* | 0.1 |
| 20 | As-F*:S-F* | As-F:S-F* | −0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlova, A.S.; Yakovleva, K.I.; Epanchitseva, A.V.; Kupryushkin, M.S.; Pyshnaya, I.A.; Pyshnyi, D.V.; Ryabchikova, E.I.; Dovydenko, I.S. An Influence of Modification with Phosphoryl Guanidine Combined with a 2′-O-Methyl or 2′-Fluoro Group on the Small-Interfering-RNA Effect. Int. J. Mol. Sci. 2021, 22, 9784. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189784
Pavlova AS, Yakovleva KI, Epanchitseva AV, Kupryushkin MS, Pyshnaya IA, Pyshnyi DV, Ryabchikova EI, Dovydenko IS. An Influence of Modification with Phosphoryl Guanidine Combined with a 2′-O-Methyl or 2′-Fluoro Group on the Small-Interfering-RNA Effect. International Journal of Molecular Sciences. 2021; 22(18):9784. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189784
Chicago/Turabian StylePavlova, Anna S., Kristina I. Yakovleva, Anna V. Epanchitseva, Maxim S. Kupryushkin, Inna A. Pyshnaya, Dmitrii V. Pyshnyi, Elena I. Ryabchikova, and Ilya S. Dovydenko. 2021. "An Influence of Modification with Phosphoryl Guanidine Combined with a 2′-O-Methyl or 2′-Fluoro Group on the Small-Interfering-RNA Effect" International Journal of Molecular Sciences 22, no. 18: 9784. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189784